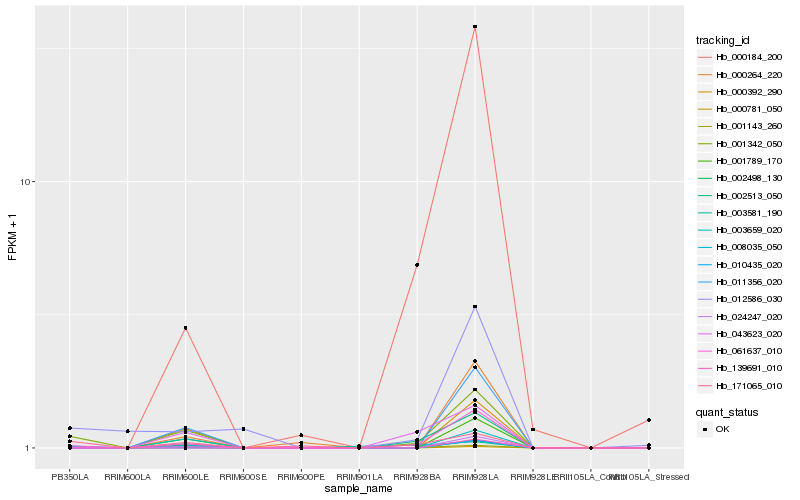
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001342_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_171065_010 |
0.2527152602 |
- |
- |
Ribonuclease H protein [Theobroma cacao] |
| 3 |
Hb_001143_260 |
0.2770748478 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 4 |
Hb_001789_170 |
0.2819831314 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |
| 5 |
Hb_002498_130 |
0.2822135497 |
- |
- |
hypothetical protein POPTR_0004s00980g [Populus trichocarpa] |
| 6 |
Hb_000781_050 |
0.2827260631 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_011356_020 |
0.2849021344 |
- |
- |
- |
| 8 |
Hb_000392_290 |
0.2871617197 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_139691_010 |
0.2999958321 |
- |
- |
- |
| 10 |
Hb_024247_020 |
0.3023388436 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 11 |
Hb_012586_030 |
0.3030711345 |
- |
- |
hypothetical protein CICLE_v10014868mg [Citrus clementina] |
| 12 |
Hb_008035_050 |
0.3033536417 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 13 |
Hb_003659_020 |
0.3051809054 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 14 |
Hb_043623_020 |
0.3133018813 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 15 |
Hb_000184_200 |
0.3242341648 |
- |
- |
- |
| 16 |
Hb_002513_050 |
0.3251343076 |
- |
- |
hypothetical protein F383_14794 [Gossypium arboreum] |
| 17 |
Hb_061637_010 |
0.3304149865 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 18 |
Hb_003581_190 |
0.3332471975 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 19 |
Hb_010435_020 |
0.3395184072 |
- |
- |
PREDICTED: uncharacterized protein LOC104216897 [Nicotiana sylvestris] |
| 20 |
Hb_000264_220 |
0.3396625928 |
- |
- |
PREDICTED: uncharacterized protein LOC105649527 [Jatropha curcas] |