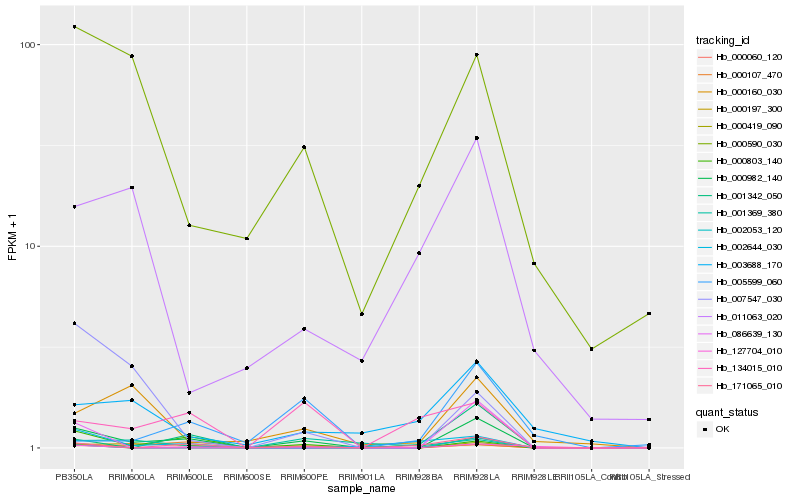
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000982_140 |
0.0 |
- |
- |
histidine kinase 1 plant, putative [Ricinus communis] |
| 2 |
Hb_000197_300 |
0.2864916447 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 3 |
Hb_000803_140 |
0.288722588 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase-like [Populus euphratica] |
| 4 |
Hb_171065_010 |
0.3106509814 |
- |
- |
Ribonuclease H protein [Theobroma cacao] |
| 5 |
Hb_000590_030 |
0.3681490386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_134015_010 |
0.3777767428 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 7 |
Hb_000060_120 |
0.3778895156 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 8 |
Hb_000160_030 |
0.3868844541 |
- |
- |
putative S-adenosylmethionine-dependent methyltransferase [Morus notabilis] |
| 9 |
Hb_001342_050 |
0.38835272 |
- |
- |
- |
| 10 |
Hb_000419_090 |
0.395013508 |
- |
- |
hypothetical protein JCGZ_18532 [Jatropha curcas] |
| 11 |
Hb_127704_010 |
0.3975982151 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_003688_170 |
0.4002834684 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 isoform X2 [Cucumis sativus] |
| 13 |
Hb_086639_130 |
0.4018624544 |
- |
- |
- |
| 14 |
Hb_002644_030 |
0.4063857511 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like, partial [Eucalyptus grandis] |
| 15 |
Hb_007547_030 |
0.4098829751 |
- |
- |
vacuolar sorting receptor family protein [Populus trichocarpa] |
| 16 |
Hb_011063_020 |
0.4126116207 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_001369_380 |
0.4138001582 |
- |
- |
F-box family protein, putative [Theobroma cacao] |
| 18 |
Hb_005599_060 |
0.4165424069 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000107_470 |
0.4167783878 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 20 |
Hb_002053_120 |
0.420064936 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |