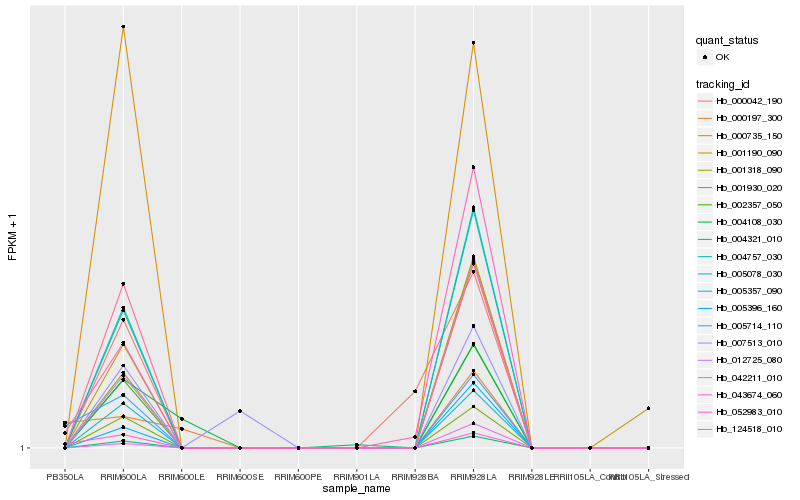
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004108_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_002357_050 |
0.2698548232 |
- |
- |
- |
| 3 |
Hb_001930_020 |
0.2717003534 |
- |
- |
PREDICTED: pectinesterase QRT1 [Jatropha curcas] |
| 4 |
Hb_005078_030 |
0.2720708588 |
- |
- |
- |
| 5 |
Hb_005357_090 |
0.2722424854 |
- |
- |
ubiquitin, putative [Ricinus communis] |
| 6 |
Hb_004757_030 |
0.2722547767 |
- |
- |
- |
| 7 |
Hb_124518_010 |
0.277100399 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 8 |
Hb_000735_150 |
0.2880672148 |
- |
- |
hypothetical protein JCGZ_15322 [Jatropha curcas] |
| 9 |
Hb_001318_090 |
0.2918221396 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 10 |
Hb_012725_080 |
0.2923453472 |
- |
- |
- |
| 11 |
Hb_005396_160 |
0.3029708782 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 12 |
Hb_000197_300 |
0.3051294295 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 13 |
Hb_052983_010 |
0.3366070811 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 14 |
Hb_042211_010 |
0.3492383229 |
- |
- |
PREDICTED: uncharacterized protein LOC103404019 [Malus domestica] |
| 15 |
Hb_001190_090 |
0.3549038749 |
- |
- |
PREDICTED: uncharacterized protein LOC105638813 [Jatropha curcas] |
| 16 |
Hb_043674_060 |
0.3696924657 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 17 |
Hb_004321_010 |
0.3766903733 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_005714_110 |
0.3769647224 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA10 [Jatropha curcas] |
| 19 |
Hb_007513_010 |
0.3771676426 |
- |
- |
- |
| 20 |
Hb_000042_190 |
0.3776726384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |