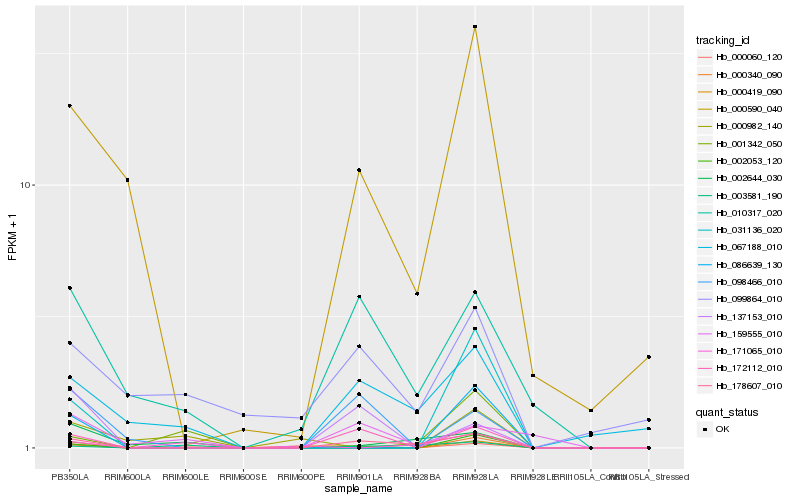
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000060_120 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 2 |
Hb_000419_090 |
0.3180106675 |
- |
- |
hypothetical protein JCGZ_18532 [Jatropha curcas] |
| 3 |
Hb_172112_010 |
0.3213532629 |
- |
- |
hypothetical protein JCGZ_26322 [Jatropha curcas] |
| 4 |
Hb_178607_010 |
0.3359874766 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 5 |
Hb_086639_130 |
0.3544908715 |
- |
- |
- |
| 6 |
Hb_002644_030 |
0.3586949833 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like, partial [Eucalyptus grandis] |
| 7 |
Hb_159555_010 |
0.3618004051 |
- |
- |
PREDICTED: uncharacterized protein LOC105795953 [Gossypium raimondii] |
| 8 |
Hb_010317_020 |
0.3626488179 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 9 |
Hb_137153_010 |
0.3676813664 |
- |
- |
- |
| 10 |
Hb_098466_010 |
0.3744602136 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 11 |
Hb_003581_190 |
0.377191837 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 12 |
Hb_000982_140 |
0.3778895156 |
- |
- |
histidine kinase 1 plant, putative [Ricinus communis] |
| 13 |
Hb_067188_010 |
0.3796603471 |
- |
- |
- |
| 14 |
Hb_000590_040 |
0.3845885055 |
- |
- |
hypothetical protein MPER_09217 [Moniliophthora perniciosa FA553] |
| 15 |
Hb_002053_120 |
0.3876719755 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |
| 16 |
Hb_031136_020 |
0.3888881034 |
- |
- |
- |
| 17 |
Hb_000340_090 |
0.3890812121 |
- |
- |
- |
| 18 |
Hb_001342_050 |
0.3943836319 |
- |
- |
- |
| 19 |
Hb_099864_010 |
0.3993846437 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 20 |
Hb_171065_010 |
0.4008256505 |
- |
- |
Ribonuclease H protein [Theobroma cacao] |