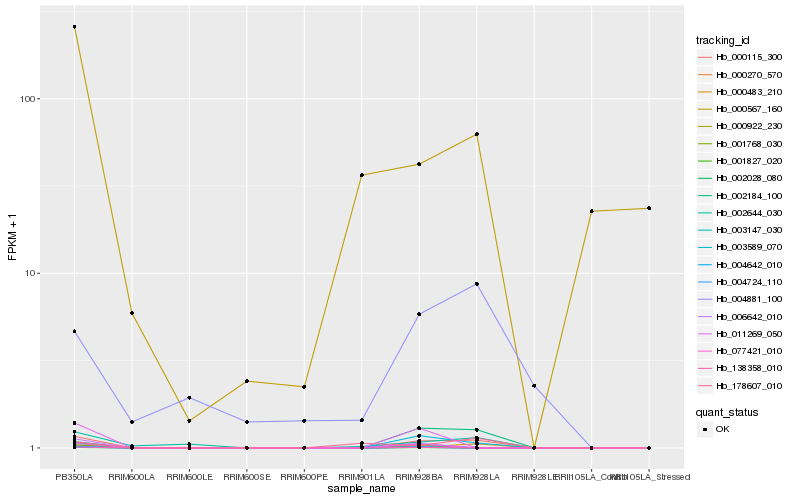
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_300 |
0.0 |
- |
- |
- |
| 2 |
Hb_000483_210 |
0.2438673978 |
- |
- |
hypothetical protein JCGZ_03497 [Jatropha curcas] |
| 3 |
Hb_003589_070 |
0.2540807195 |
- |
- |
PREDICTED: uncharacterized protein LOC104242373 [Nicotiana sylvestris] |
| 4 |
Hb_002184_100 |
0.3002166361 |
- |
- |
hypothetical protein PRUPE_ppa020120mg [Prunus persica] |
| 5 |
Hb_002644_030 |
0.319129252 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like, partial [Eucalyptus grandis] |
| 6 |
Hb_004881_100 |
0.3782366631 |
- |
- |
Phosphoribosyltransferase family protein isoform 4 [Theobroma cacao] |
| 7 |
Hb_178607_010 |
0.3928664153 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 8 |
Hb_000567_160 |
0.3945422823 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Jatropha curcas] |
| 9 |
Hb_006642_010 |
0.4056091601 |
- |
- |
- |
| 10 |
Hb_003147_030 |
0.4059430045 |
transcription factor |
TF Family: Jumonji |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_138358_010 |
0.4059607131 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 12 |
Hb_004642_010 |
0.40598842 |
- |
- |
Integrase core domain, putative [Oryza sativa Japonica Group] |
| 13 |
Hb_000270_570 |
0.4061906014 |
- |
- |
PREDICTED: peroxidase 65-like [Jatropha curcas] |
| 14 |
Hb_004724_110 |
0.4062732236 |
- |
- |
alpha-1,2-fucosyltransferase [Populus tremula x Populus alba] |
| 15 |
Hb_000922_230 |
0.4065669261 |
- |
- |
- |
| 16 |
Hb_001768_030 |
0.4067308515 |
- |
- |
- |
| 17 |
Hb_001827_020 |
0.4071186739 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_011269_050 |
0.4071990098 |
- |
- |
- |
| 19 |
Hb_077421_010 |
0.4073988135 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 20 |
Hb_002028_080 |
0.4074221153 |
- |
- |
hypothetical protein JCGZ_07571 [Jatropha curcas] |