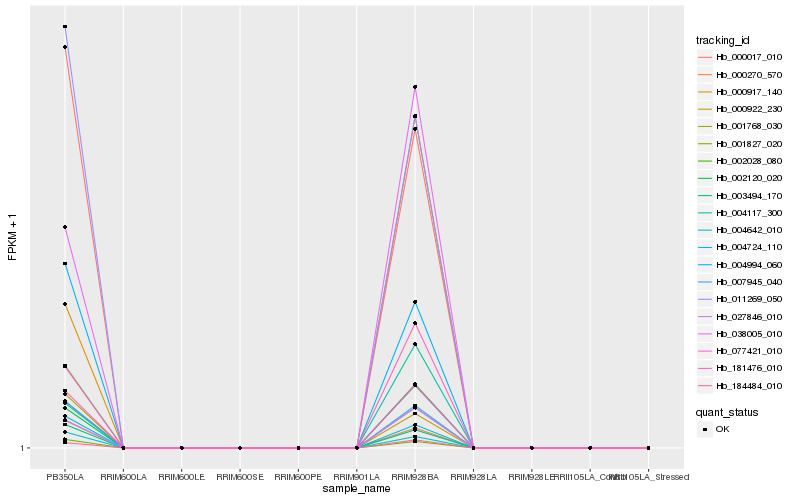
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038005_010 |
0.0 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 2 |
Hb_002120_020 |
0.0199777438 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 3 |
Hb_184484_010 |
0.0284882572 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_027846_010 |
0.0375768285 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 5 |
Hb_181476_010 |
0.0513221797 |
- |
- |
- |
| 6 |
Hb_004117_300 |
0.0626398977 |
- |
- |
- |
| 7 |
Hb_000917_140 |
0.0713107227 |
- |
- |
- |
| 8 |
Hb_007945_040 |
0.1367275799 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 9 |
Hb_004994_060 |
0.1673158683 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 10 |
Hb_003494_170 |
0.1698556237 |
- |
- |
- |
| 11 |
Hb_000017_010 |
0.1702156722 |
- |
- |
- |
| 12 |
Hb_002028_080 |
0.1706119144 |
- |
- |
hypothetical protein JCGZ_07571 [Jatropha curcas] |
| 13 |
Hb_077421_010 |
0.1708259031 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 14 |
Hb_011269_050 |
0.1726970199 |
- |
- |
- |
| 15 |
Hb_001827_020 |
0.1734686915 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_001768_030 |
0.1773719677 |
- |
- |
- |
| 17 |
Hb_000922_230 |
0.1791233032 |
- |
- |
- |
| 18 |
Hb_004724_110 |
0.1824451285 |
- |
- |
alpha-1,2-fucosyltransferase [Populus tremula x Populus alba] |
| 19 |
Hb_000270_570 |
0.1834286544 |
- |
- |
PREDICTED: peroxidase 65-like [Jatropha curcas] |
| 20 |
Hb_004642_010 |
0.1859429 |
- |
- |
Integrase core domain, putative [Oryza sativa Japonica Group] |