| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
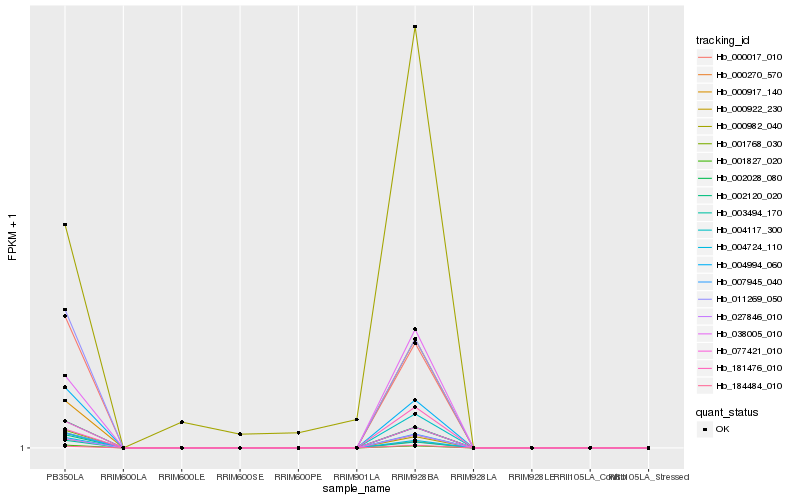
Hb_181476_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004117_300 |
0.01133394 |
- |
- |
- |
| 3 |
Hb_000917_140 |
0.0200214679 |
- |
- |
- |
| 4 |
Hb_038005_010 |
0.0513221797 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 5 |
Hb_002120_020 |
0.0712698928 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 6 |
Hb_184484_010 |
0.0797628648 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_027846_010 |
0.0888296666 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 8 |
Hb_007945_040 |
0.1875543842 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 9 |
Hb_000982_040 |
0.2002153026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004994_060 |
0.2179460758 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 11 |
Hb_003494_170 |
0.2204681795 |
- |
- |
- |
| 12 |
Hb_000017_010 |
0.2208257092 |
- |
- |
- |
| 13 |
Hb_002028_080 |
0.2212191746 |
- |
- |
hypothetical protein JCGZ_07571 [Jatropha curcas] |
| 14 |
Hb_077421_010 |
0.2214316617 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 15 |
Hb_011269_050 |
0.2232895869 |
- |
- |
- |
| 16 |
Hb_001827_020 |
0.2240557861 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_001768_030 |
0.2279310948 |
- |
- |
- |
| 18 |
Hb_000922_230 |
0.2296697261 |
- |
- |
- |
| 19 |
Hb_004724_110 |
0.2329671901 |
- |
- |
alpha-1,2-fucosyltransferase [Populus tremula x Populus alba] |
| 20 |
Hb_000270_570 |
0.2339434365 |
- |
- |
PREDICTED: peroxidase 65-like [Jatropha curcas] |