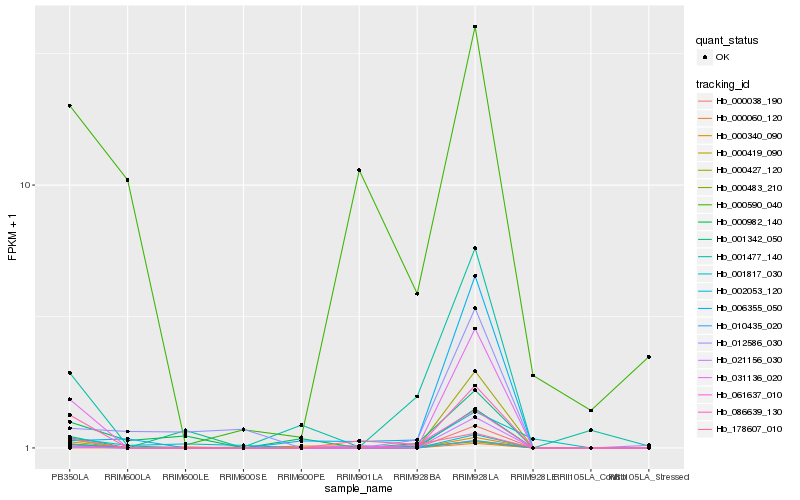
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086639_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_002053_120 |
0.0850340895 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |
| 3 |
Hb_031136_020 |
0.0876187679 |
- |
- |
- |
| 4 |
Hb_000340_090 |
0.0880270557 |
- |
- |
- |
| 5 |
Hb_010435_020 |
0.1924773558 |
- |
- |
PREDICTED: uncharacterized protein LOC104216897 [Nicotiana sylvestris] |
| 6 |
Hb_000483_210 |
0.2545067264 |
- |
- |
hypothetical protein JCGZ_03497 [Jatropha curcas] |
| 7 |
Hb_000427_120 |
0.2678927647 |
- |
- |
- |
| 8 |
Hb_001477_140 |
0.3174101637 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000060_120 |
0.3544908715 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 10 |
Hb_061637_010 |
0.3568893107 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 11 |
Hb_000419_090 |
0.3645448457 |
- |
- |
hypothetical protein JCGZ_18532 [Jatropha curcas] |
| 12 |
Hb_021156_030 |
0.3731716238 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 13 |
Hb_001342_050 |
0.3790659262 |
- |
- |
- |
| 14 |
Hb_000982_140 |
0.4018624544 |
- |
- |
histidine kinase 1 plant, putative [Ricinus communis] |
| 15 |
Hb_012586_030 |
0.4023623985 |
- |
- |
hypothetical protein CICLE_v10014868mg [Citrus clementina] |
| 16 |
Hb_006355_050 |
0.4058045857 |
- |
- |
- |
| 17 |
Hb_001817_030 |
0.4095777495 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 18 |
Hb_000590_040 |
0.4152429791 |
- |
- |
hypothetical protein MPER_09217 [Moniliophthora perniciosa FA553] |
| 19 |
Hb_178607_010 |
0.416455815 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 20 |
Hb_000038_190 |
0.4226301695 |
- |
- |
- |