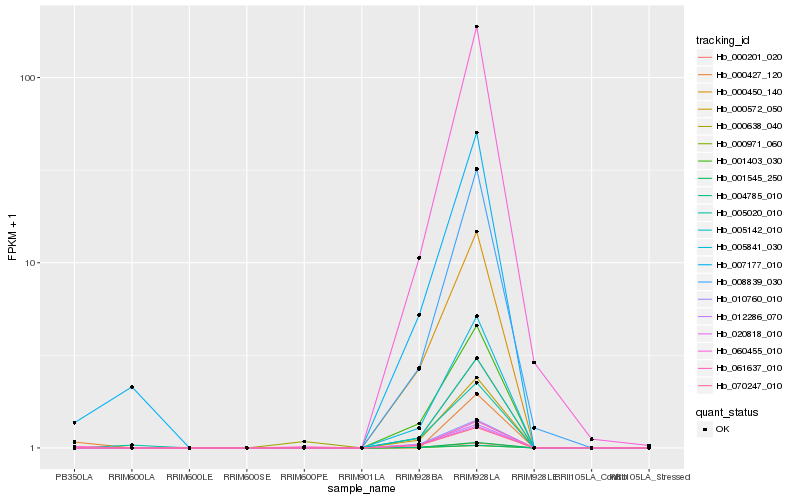
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061637_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 2 |
Hb_007177_010 |
0.1446935646 |
- |
- |
- |
| 3 |
Hb_000572_050 |
0.1458652315 |
- |
- |
PREDICTED: transcriptional adapter ADA2b isoform X1 [Cicer arietinum] |
| 4 |
Hb_005841_030 |
0.1460189853 |
- |
- |
- |
| 5 |
Hb_005142_010 |
0.1462808875 |
- |
- |
- |
| 6 |
Hb_001403_030 |
0.1501051461 |
- |
- |
- |
| 7 |
Hb_000971_060 |
0.151310223 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000201_020 |
0.1518702165 |
- |
- |
hypothetical protein CICLE_v10024145mg [Citrus clementina] |
| 9 |
Hb_001545_250 |
0.1519813303 |
- |
- |
PREDICTED: uncharacterized protein LOC103936764 [Pyrus x bretschneideri] |
| 10 |
Hb_010760_010 |
0.1547039273 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 11 |
Hb_070247_010 |
0.156688218 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 12 |
Hb_020818_010 |
0.1572700014 |
- |
- |
PREDICTED: uncharacterized protein LOC105648455 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000450_140 |
0.1581793593 |
- |
- |
hypothetical protein POPTR_0009s06810g [Populus trichocarpa] |
| 14 |
Hb_012286_070 |
0.1582171246 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 15 |
Hb_008839_030 |
0.1617561304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_060455_010 |
0.1655597911 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 17 |
Hb_005020_010 |
0.1872806665 |
- |
- |
- |
| 18 |
Hb_000427_120 |
0.1885366718 |
- |
- |
- |
| 19 |
Hb_000638_040 |
0.1986604308 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-7-like [Glycine max] |
| 20 |
Hb_004785_010 |
0.1995721512 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |