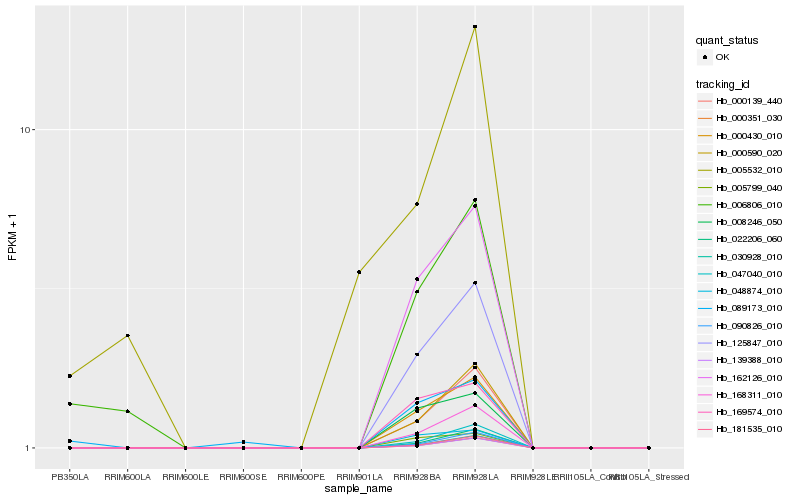
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006806_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_089173_010 |
0.2089165621 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 3 |
Hb_125847_010 |
0.2121987609 |
- |
- |
- |
| 4 |
Hb_000430_010 |
0.2128281075 |
- |
- |
- |
| 5 |
Hb_162126_010 |
0.2151032185 |
- |
- |
- |
| 6 |
Hb_168311_010 |
0.21930697 |
- |
- |
- |
| 7 |
Hb_000139_440 |
0.2259353806 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_000590_020 |
0.2297725997 |
- |
- |
hypothetical protein JCGZ_18748 [Jatropha curcas] |
| 9 |
Hb_047040_010 |
0.2315445614 |
- |
- |
integrase [Gossypium herbaceum] |
| 10 |
Hb_008246_050 |
0.2332332462 |
- |
- |
- |
| 11 |
Hb_005799_040 |
0.2332518903 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |
| 12 |
Hb_022206_060 |
0.2348927148 |
- |
- |
- |
| 13 |
Hb_048874_010 |
0.2358515997 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |
| 14 |
Hb_090826_010 |
0.2367499275 |
- |
- |
PREDICTED: uncharacterized protein LOC105786910 [Gossypium raimondii] |
| 15 |
Hb_169574_010 |
0.2369421279 |
- |
- |
PREDICTED: uncharacterized protein LOC104120373 [Nicotiana tomentosiformis] |
| 16 |
Hb_005532_010 |
0.2376130751 |
- |
- |
- |
| 17 |
Hb_030928_010 |
0.23882594 |
- |
- |
- |
| 18 |
Hb_181535_010 |
0.239137664 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_000351_030 |
0.2402099329 |
- |
- |
putative NBS-LRR type disease resistance protein [Prunus persica] |
| 20 |
Hb_139388_010 |
0.240354118 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |