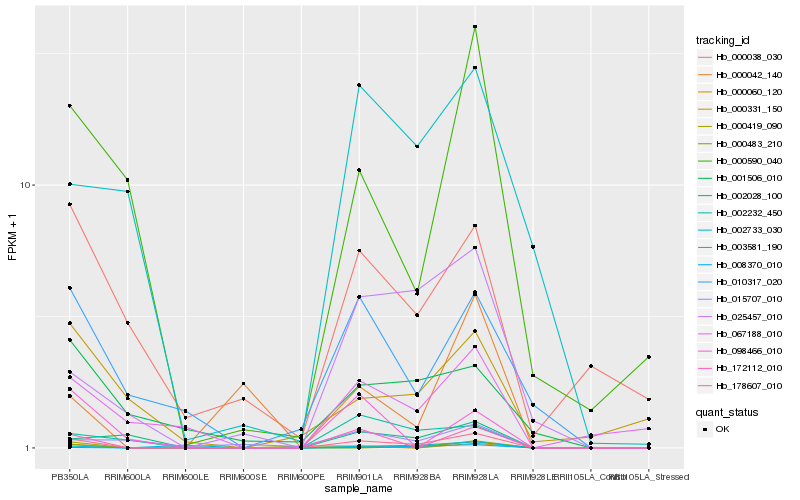
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178607_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 2 |
Hb_172112_010 |
0.2586142504 |
- |
- |
hypothetical protein JCGZ_26322 [Jatropha curcas] |
| 3 |
Hb_025457_010 |
0.304511399 |
- |
- |
- |
| 4 |
Hb_000590_040 |
0.3062730258 |
- |
- |
hypothetical protein MPER_09217 [Moniliophthora perniciosa FA553] |
| 5 |
Hb_010317_020 |
0.3099944132 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 6 |
Hb_067188_010 |
0.3111033161 |
- |
- |
- |
| 7 |
Hb_002232_450 |
0.3133102112 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 8 |
Hb_008370_010 |
0.3147536946 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_000483_210 |
0.32991142 |
- |
- |
hypothetical protein JCGZ_03497 [Jatropha curcas] |
| 10 |
Hb_002028_100 |
0.333052427 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 11 |
Hb_000060_120 |
0.3359874766 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 12 |
Hb_000038_030 |
0.3373172256 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |
| 13 |
Hb_001506_010 |
0.3411453535 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 14 |
Hb_000042_140 |
0.3418816368 |
- |
- |
hypothetical protein POPTR_0001s41640g, partial [Populus trichocarpa] |
| 15 |
Hb_000419_090 |
0.3444154281 |
- |
- |
hypothetical protein JCGZ_18532 [Jatropha curcas] |
| 16 |
Hb_002733_030 |
0.3451316721 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 17 |
Hb_098466_010 |
0.3475865163 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 18 |
Hb_000331_150 |
0.3512953377 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_015707_010 |
0.3527166962 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 20 |
Hb_003581_190 |
0.3581958067 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_21694 [Jatropha curcas] |