| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
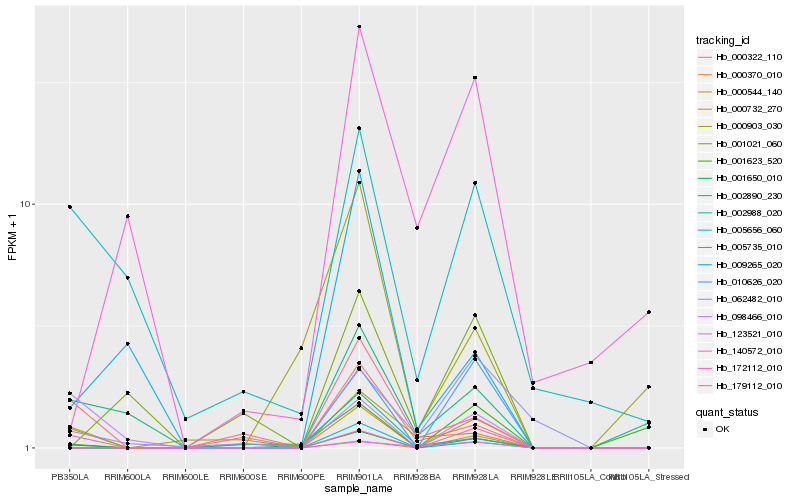
Hb_001650_010 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_172112_010 |
0.2187156191 |
- |
- |
hypothetical protein JCGZ_26322 [Jatropha curcas] |
| 3 |
Hb_000544_140 |
0.2279625491 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 4 |
Hb_010626_020 |
0.2723839499 |
- |
- |
- |
| 5 |
Hb_005735_010 |
0.2778665953 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_005656_060 |
0.3286893392 |
- |
- |
- |
| 7 |
Hb_002890_230 |
0.3288171112 |
- |
- |
hypothetical protein MTR_5g084920 [Medicago truncatula] |
| 8 |
Hb_123521_010 |
0.329297034 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 9 |
Hb_000370_010 |
0.3309510601 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 10 |
Hb_000322_110 |
0.3343627299 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 3 [Jatropha curcas] |
| 11 |
Hb_062482_010 |
0.3419753366 |
- |
- |
- |
| 12 |
Hb_098466_010 |
0.3443364305 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 13 |
Hb_179112_010 |
0.3567114089 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 14 |
Hb_002988_020 |
0.3568971479 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 15 |
Hb_009265_020 |
0.3625341492 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001623_520 |
0.3692097314 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 17 |
Hb_000903_030 |
0.3721482214 |
- |
- |
- |
| 18 |
Hb_001021_060 |
0.3743474395 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000732_270 |
0.3771969868 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 20 |
Hb_140572_010 |
0.3780057214 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |