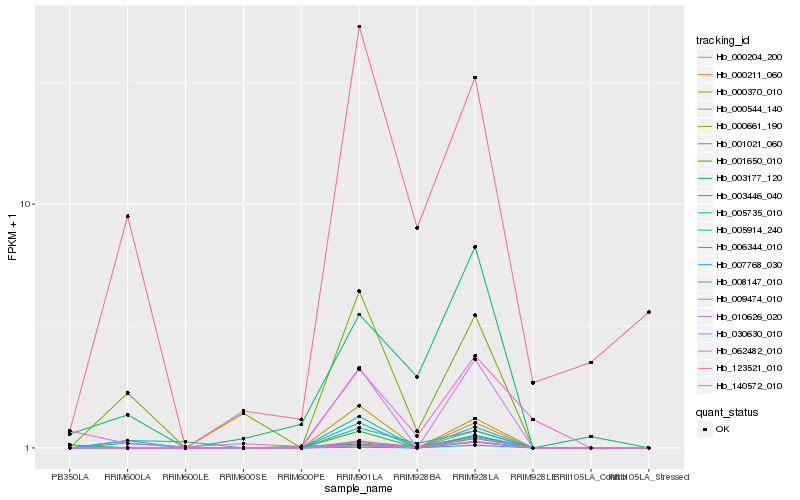
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010626_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000370_010 |
0.0927551912 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 3 |
Hb_000544_140 |
0.1201684863 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 4 |
Hb_030630_010 |
0.1970895328 |
- |
- |
PREDICTED: uncharacterized protein LOC105786776 [Gossypium raimondii] |
| 5 |
Hb_000204_200 |
0.2017015944 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 6 |
Hb_123521_010 |
0.2255903531 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 7 |
Hb_009474_010 |
0.22603776 |
- |
- |
- |
| 8 |
Hb_007768_030 |
0.2353170091 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_001650_010 |
0.2723839499 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 10 |
Hb_003177_120 |
0.2880140546 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Populus euphratica] |
| 11 |
Hb_000661_190 |
0.2919355513 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 12 |
Hb_001021_060 |
0.3181632529 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_005735_010 |
0.3213325893 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_005914_240 |
0.3414348625 |
- |
- |
PREDICTED: oxidation resistance protein 1 [Jatropha curcas] |
| 15 |
Hb_003446_040 |
0.3437949179 |
- |
- |
- |
| 16 |
Hb_000211_060 |
0.3448492755 |
- |
- |
glutathione peroxidase [Populus euphratica] |
| 17 |
Hb_062482_010 |
0.3480138936 |
- |
- |
- |
| 18 |
Hb_140572_010 |
0.3484452982 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 19 |
Hb_006344_010 |
0.3610076499 |
- |
- |
- |
| 20 |
Hb_008147_010 |
0.362512425 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |