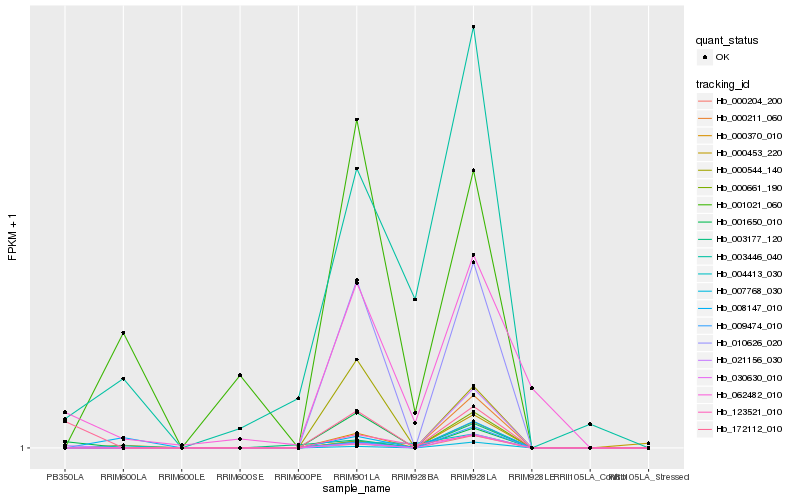
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000370_010 |
0.0 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 2 |
Hb_010626_020 |
0.0927551912 |
- |
- |
- |
| 3 |
Hb_030630_010 |
0.1051298005 |
- |
- |
PREDICTED: uncharacterized protein LOC105786776 [Gossypium raimondii] |
| 4 |
Hb_000204_200 |
0.1098002174 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 5 |
Hb_009474_010 |
0.1344816295 |
- |
- |
- |
| 6 |
Hb_007768_030 |
0.1439097836 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_000661_190 |
0.201667052 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 8 |
Hb_000544_140 |
0.2120992973 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 9 |
Hb_003177_120 |
0.2360581875 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Populus euphratica] |
| 10 |
Hb_000211_060 |
0.2560615413 |
- |
- |
glutathione peroxidase [Populus euphratica] |
| 11 |
Hb_123521_010 |
0.2577833114 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 12 |
Hb_003446_040 |
0.3214899031 |
- |
- |
- |
| 13 |
Hb_001650_010 |
0.3309510601 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 14 |
Hb_021156_030 |
0.3377094009 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 15 |
Hb_008147_010 |
0.3395425529 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 16 |
Hb_001021_060 |
0.3536043421 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_062482_010 |
0.3548752149 |
- |
- |
- |
| 18 |
Hb_000453_220 |
0.3645800774 |
- |
- |
hypothetical protein JCGZ_11079 [Jatropha curcas] |
| 19 |
Hb_004413_030 |
0.374582683 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 20 |
Hb_172112_010 |
0.3791753004 |
- |
- |
hypothetical protein JCGZ_26322 [Jatropha curcas] |