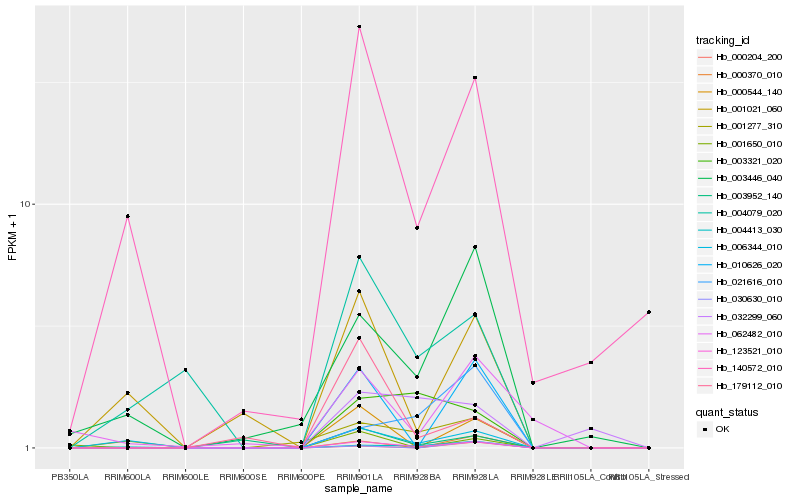
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123521_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 2 |
Hb_010626_020 |
0.2255903531 |
- |
- |
- |
| 3 |
Hb_000544_140 |
0.2345590241 |
- |
- |
PREDICTED: uncharacterized protein LOC101245866 [Solanum lycopersicum] |
| 4 |
Hb_000370_010 |
0.2577833114 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 5 |
Hb_140572_010 |
0.2749303068 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 6 |
Hb_003446_040 |
0.2762956794 |
- |
- |
- |
| 7 |
Hb_006344_010 |
0.2775348019 |
- |
- |
- |
| 8 |
Hb_004413_030 |
0.2906413988 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 9 |
Hb_001277_310 |
0.3054056706 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Phoenix dactylifera] |
| 10 |
Hb_001021_060 |
0.308136521 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_004079_020 |
0.3098193252 |
- |
- |
hypothetical protein glysoja_031528 [Glycine soja] |
| 12 |
Hb_003321_020 |
0.315234195 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 13 |
Hb_030630_010 |
0.3215072383 |
- |
- |
PREDICTED: uncharacterized protein LOC105786776 [Gossypium raimondii] |
| 14 |
Hb_003952_140 |
0.3222046891 |
- |
- |
hypothetical protein POPTR_0001s24960g [Populus trichocarpa] |
| 15 |
Hb_000204_200 |
0.3247648701 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 16 |
Hb_062482_010 |
0.3287442487 |
- |
- |
- |
| 17 |
Hb_001650_010 |
0.329297034 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 18 |
Hb_021616_010 |
0.3329809792 |
- |
- |
- |
| 19 |
Hb_032299_060 |
0.3414027978 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Elaeis guineensis] |
| 20 |
Hb_179112_010 |
0.3421961958 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |