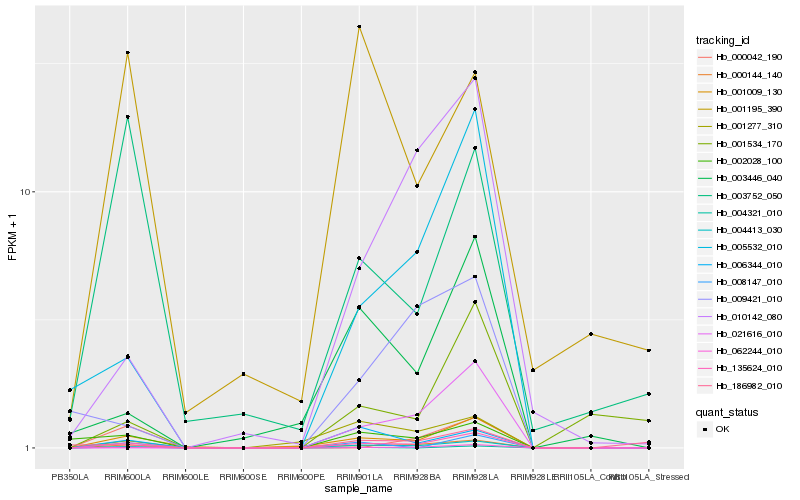
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135624_010 |
0.0 |
- |
- |
PREDICTED: inositol transporter 4-like [Jatropha curcas] |
| 2 |
Hb_062244_010 |
0.1220945317 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_004413_030 |
0.1350343254 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 4 |
Hb_001009_130 |
0.1418734005 |
- |
- |
hypothetical protein JCGZ_11336 [Jatropha curcas] |
| 5 |
Hb_005532_010 |
0.235360991 |
- |
- |
- |
| 6 |
Hb_000144_140 |
0.249704809 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 7 |
Hb_006344_010 |
0.2535404402 |
- |
- |
- |
| 8 |
Hb_008147_010 |
0.2687941446 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 9 |
Hb_002028_100 |
0.2719802516 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 10 |
Hb_003446_040 |
0.2728928389 |
- |
- |
- |
| 11 |
Hb_001277_310 |
0.2816324444 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Phoenix dactylifera] |
| 12 |
Hb_010142_080 |
0.2830445227 |
- |
- |
PREDICTED: uncharacterized protein LOC105630660 [Jatropha curcas] |
| 13 |
Hb_004321_010 |
0.2832928408 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_021616_010 |
0.3137993559 |
- |
- |
- |
| 15 |
Hb_000042_190 |
0.3169891486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001534_170 |
0.3226116423 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |
| 17 |
Hb_009421_010 |
0.3241874088 |
- |
- |
- |
| 18 |
Hb_003752_050 |
0.3245883083 |
- |
- |
hypothetical protein JCGZ_15078 [Jatropha curcas] |
| 19 |
Hb_186982_010 |
0.3375443625 |
- |
- |
PREDICTED: uncharacterized protein LOC104120222 [Nicotiana tomentosiformis] |
| 20 |
Hb_001195_390 |
0.3410087495 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Populus euphratica] |