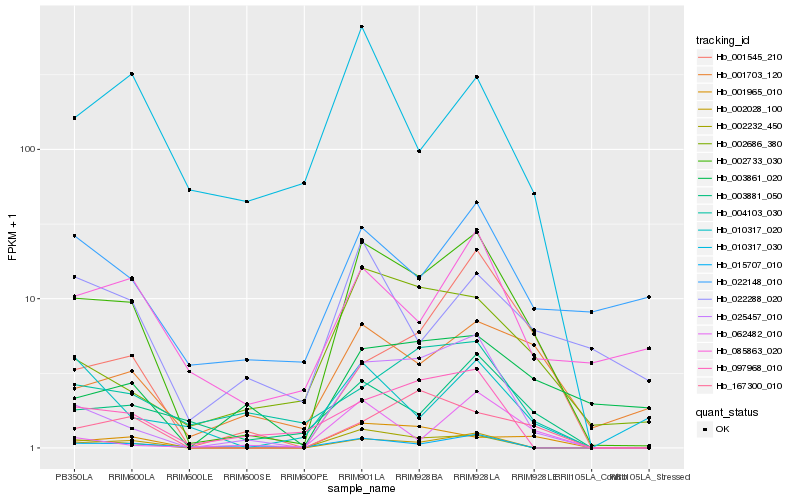
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002733_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 2 |
Hb_025457_010 |
0.1926758637 |
- |
- |
- |
| 3 |
Hb_002028_100 |
0.2060206707 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 4 |
Hb_002232_450 |
0.2102626846 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 5 |
Hb_003881_050 |
0.2401937982 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 6 |
Hb_001965_010 |
0.2466012776 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 7 |
Hb_002686_380 |
0.2481382205 |
- |
- |
- |
| 8 |
Hb_001703_120 |
0.2594293019 |
- |
- |
PREDICTED: uncharacterized protein LOC105116966 isoform X2 [Populus euphratica] |
| 9 |
Hb_015707_010 |
0.2625360167 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 10 |
Hb_167300_010 |
0.2756831593 |
- |
- |
- |
| 11 |
Hb_010317_020 |
0.2783590999 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 12 |
Hb_085863_020 |
0.2785248836 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 13 |
Hb_097968_010 |
0.2798594504 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 14 |
Hb_062482_010 |
0.2812164887 |
- |
- |
- |
| 15 |
Hb_001545_210 |
0.2852629004 |
- |
- |
hypothetical protein JCGZ_11132 [Jatropha curcas] |
| 16 |
Hb_003861_020 |
0.2890397642 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like isoform X2 [Nelumbo nucifera] |
| 17 |
Hb_004103_030 |
0.2897339085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_022288_020 |
0.2930398212 |
- |
- |
hypothetical protein POPTR_0006s08590g [Populus trichocarpa] |
| 19 |
Hb_010317_030 |
0.2947194763 |
- |
- |
unknown [Picea sitchensis] |
| 20 |
Hb_022148_010 |
0.2956068208 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |