| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
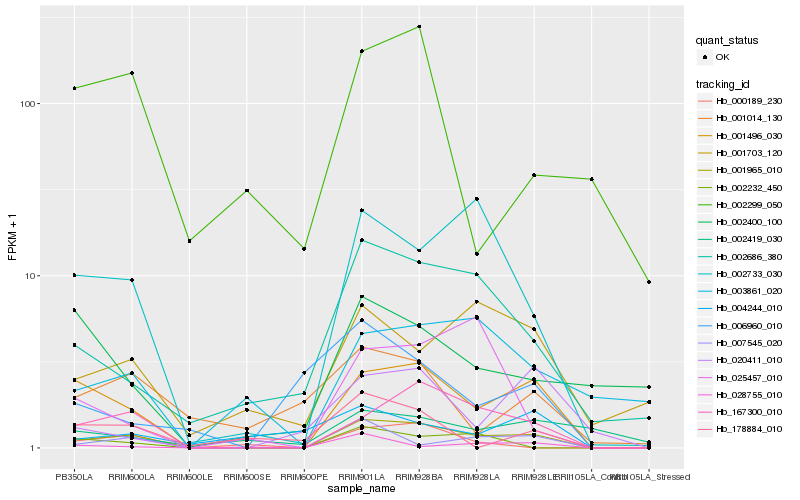
Hb_001965_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 2 |
Hb_001496_030 |
0.176857974 |
- |
- |
hypothetical protein CISIN_1g0170981mg, partial [Citrus sinensis] |
| 3 |
Hb_167300_010 |
0.2068618249 |
- |
- |
- |
| 4 |
Hb_002733_030 |
0.2466012776 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 5 |
Hb_002686_380 |
0.2682675983 |
- |
- |
- |
| 6 |
Hb_007545_020 |
0.2914159679 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Nicotiana tomentosiformis] |
| 7 |
Hb_001703_120 |
0.3088855807 |
- |
- |
PREDICTED: uncharacterized protein LOC105116966 isoform X2 [Populus euphratica] |
| 8 |
Hb_002232_450 |
0.3131656674 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 9 |
Hb_000189_230 |
0.3135816646 |
- |
- |
Auxin-induced protein [Aegilops tauschii] |
| 10 |
Hb_003861_020 |
0.3205065232 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like isoform X2 [Nelumbo nucifera] |
| 11 |
Hb_020411_010 |
0.3270407956 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_178884_010 |
0.3369607964 |
- |
- |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 13 |
Hb_001014_130 |
0.3422157911 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 14 |
Hb_006960_010 |
0.3433308765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002400_100 |
0.3440568975 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 16 |
Hb_028755_010 |
0.3450087625 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_025457_010 |
0.3450419294 |
- |
- |
- |
| 18 |
Hb_004244_010 |
0.3452194321 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 19 |
Hb_002419_030 |
0.3467793044 |
- |
- |
- |
| 20 |
Hb_002299_050 |
0.3497507428 |
- |
- |
PREDICTED: chalcone--flavonone isomerase isoform X1 [Pyrus x bretschneideri] |