| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
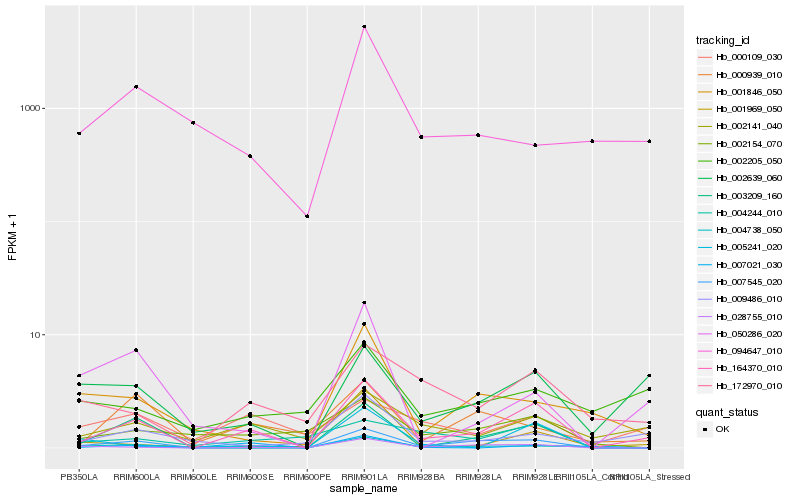
Hb_164370_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_007545_020 |
0.2784513527 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Nicotiana tomentosiformis] |
| 3 |
Hb_003209_160 |
0.2832898593 |
- |
- |
PREDICTED: putative SNAP25 homologous protein SNAP30 [Populus euphratica] |
| 4 |
Hb_002141_040 |
0.2835922178 |
- |
- |
- |
| 5 |
Hb_028755_010 |
0.2919999817 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_004738_050 |
0.3023203276 |
- |
- |
F-box/leucine rich repeat protein, putative [Ricinus communis] |
| 7 |
Hb_002639_060 |
0.3035150709 |
- |
- |
- |
| 8 |
Hb_172970_010 |
0.3089591039 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 9 |
Hb_009486_010 |
0.3146639865 |
- |
- |
- |
| 10 |
Hb_000939_010 |
0.3286074452 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 11 |
Hb_005241_020 |
0.3338807068 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 12 |
Hb_001969_050 |
0.3338854461 |
- |
- |
PREDICTED: uncharacterized protein LOC105634920 isoform X1 [Jatropha curcas] |
| 13 |
Hb_050286_020 |
0.3349855823 |
- |
- |
- |
| 14 |
Hb_000109_030 |
0.3373261641 |
- |
- |
- |
| 15 |
Hb_002154_070 |
0.3376111898 |
desease resistance |
Gene Name: AAA_25 |
hypothetical protein Csa_6G014750 [Cucumis sativus] |
| 16 |
Hb_002205_050 |
0.346559053 |
- |
- |
hypothetical protein JCGZ_03335 [Jatropha curcas] |
| 17 |
Hb_001846_050 |
0.3479448897 |
- |
- |
- |
| 18 |
Hb_004244_010 |
0.3515707672 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 19 |
Hb_094647_010 |
0.3517746453 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B-like [Jatropha curcas] |
| 20 |
Hb_007021_030 |
0.35484301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |