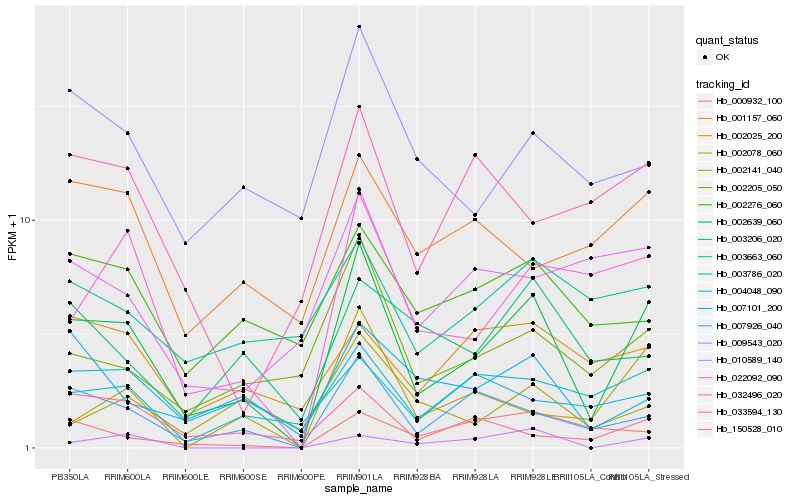
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_007926_040 |
0.1982327296 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 3 |
Hb_032496_020 |
0.2177225391 |
- |
- |
- |
| 4 |
Hb_002205_050 |
0.2298921187 |
- |
- |
hypothetical protein JCGZ_03335 [Jatropha curcas] |
| 5 |
Hb_007101_200 |
0.2456858269 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009543_020 |
0.2462858956 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 7 |
Hb_003786_020 |
0.2491934586 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 8 |
Hb_002078_060 |
0.2541259652 |
- |
- |
hypothetical protein JCGZ_17943 [Jatropha curcas] |
| 9 |
Hb_004048_090 |
0.2560584929 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g53330 [Jatropha curcas] |
| 10 |
Hb_002276_060 |
0.2584495573 |
- |
- |
hypothetical protein CISIN_1g002019mg [Citrus sinensis] |
| 11 |
Hb_002025_200 |
0.2615160093 |
- |
- |
- |
| 12 |
Hb_001157_060 |
0.2619014963 |
- |
- |
PREDICTED: putative nitric oxide synthase [Jatropha curcas] |
| 13 |
Hb_150528_010 |
0.2628582075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002141_040 |
0.2634364423 |
- |
- |
- |
| 15 |
Hb_000932_100 |
0.263705535 |
- |
- |
- |
| 16 |
Hb_003206_020 |
0.2642861679 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 17 |
Hb_003663_060 |
0.2651787296 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g43820 [Jatropha curcas] |
| 18 |
Hb_022092_090 |
0.2662175126 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 19 |
Hb_033594_130 |
0.2668503492 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Jatropha curcas] |
| 20 |
Hb_010589_140 |
0.2689088049 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Fragaria vesca subsp. vesca] |