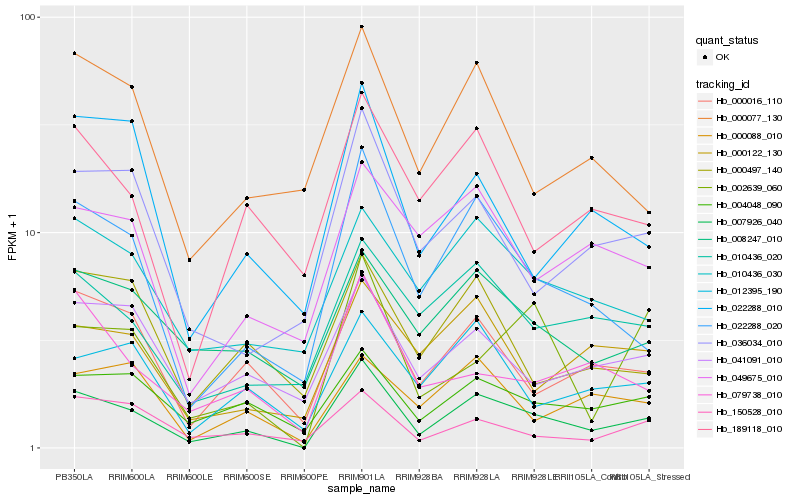
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007926_040 |
0.0 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 2 |
Hb_022288_020 |
0.1502157345 |
- |
- |
hypothetical protein POPTR_0006s08590g [Populus trichocarpa] |
| 3 |
Hb_000016_110 |
0.1672275225 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 4 |
Hb_004048_090 |
0.1754583037 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g53330 [Jatropha curcas] |
| 5 |
Hb_041091_010 |
0.1885593731 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 6 |
Hb_012395_190 |
0.1889709257 |
- |
- |
hypothetical protein B456_005G053800 [Gossypium raimondii] |
| 7 |
Hb_010436_020 |
0.1892769412 |
- |
- |
Dicer-like protein 4 [Morus notabilis] |
| 8 |
Hb_008247_010 |
0.1915421852 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 9 |
Hb_079738_010 |
0.1917065426 |
- |
- |
hypothetical protein CISIN_1g036582mg, partial [Citrus sinensis] |
| 10 |
Hb_036034_010 |
0.191757963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010436_030 |
0.1927890214 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 12 |
Hb_189118_010 |
0.1969413746 |
- |
- |
PREDICTED: extended synaptotagmin-1 [Cucumis melo] |
| 13 |
Hb_002639_060 |
0.1982327296 |
- |
- |
- |
| 14 |
Hb_150528_010 |
0.1982422948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_022288_010 |
0.1991821683 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 16 |
Hb_000122_130 |
0.2000354012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000088_010 |
0.2019380845 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_049675_010 |
0.2020595836 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 19 |
Hb_000077_130 |
0.2025743081 |
- |
- |
PREDICTED: callose synthase 12 [Jatropha curcas] |
| 20 |
Hb_000497_140 |
0.2025977742 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |