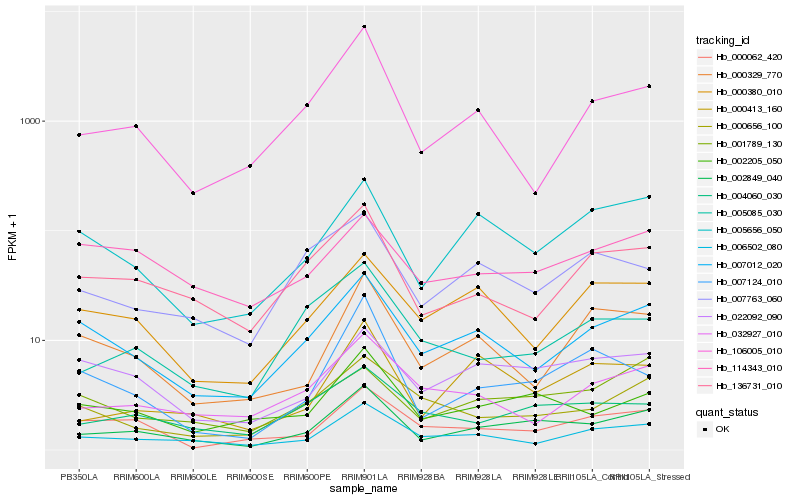
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002849_040 |
0.0 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 2 |
Hb_002205_050 |
0.14021596 |
- |
- |
hypothetical protein JCGZ_03335 [Jatropha curcas] |
| 3 |
Hb_004060_030 |
0.162449716 |
- |
- |
PREDICTED: primase homolog protein [Jatropha curcas] |
| 4 |
Hb_022092_090 |
0.163288748 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 5 |
Hb_006502_080 |
0.1679668696 |
- |
- |
- |
| 6 |
Hb_005656_050 |
0.1691509867 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 7 |
Hb_000413_160 |
0.172039827 |
- |
- |
PREDICTED: uncharacterized protein LOC105649406 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007012_020 |
0.1727192871 |
- |
- |
hypothetical protein POPTR_0005s11940g [Populus trichocarpa] |
| 9 |
Hb_136731_010 |
0.1785686036 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X4 [Jatropha curcas] |
| 10 |
Hb_001789_130 |
0.1831268829 |
- |
- |
short-chain dehydrogenase Tic32 family protein [Populus trichocarpa] |
| 11 |
Hb_000062_420 |
0.1877379294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007763_060 |
0.1906729321 |
- |
- |
BnaA02g23510D [Brassica napus] |
| 13 |
Hb_000656_100 |
0.1910053761 |
- |
- |
PREDICTED: uncharacterized protein LOC105633651 [Jatropha curcas] |
| 14 |
Hb_005085_030 |
0.1953784628 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_032927_010 |
0.1979827205 |
- |
- |
- |
| 16 |
Hb_114343_010 |
0.1982035259 |
- |
- |
PREDICTED: uncharacterized protein LOC105647831 [Jatropha curcas] |
| 17 |
Hb_000380_010 |
0.1995271313 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X3 [Jatropha curcas] |
| 18 |
Hb_007124_010 |
0.2001862582 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_000329_770 |
0.2011196006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_106005_010 |
0.2021182021 |
- |
- |
EG5651 [Manihot esculenta] |