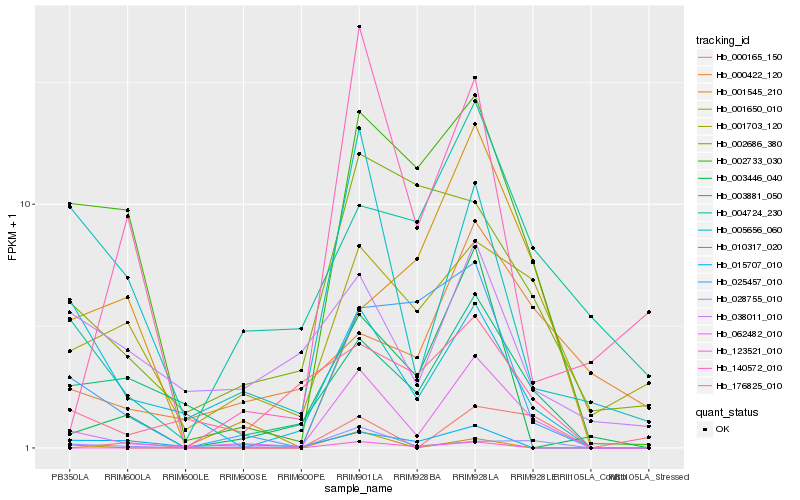
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_062482_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002733_030 |
0.2812164887 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 3 |
Hb_028755_010 |
0.2834730727 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_003881_050 |
0.2834818629 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 5 |
Hb_004724_230 |
0.3025801519 |
- |
- |
hypothetical protein JCGZ_12701 [Jatropha curcas] |
| 6 |
Hb_003446_040 |
0.3097608403 |
- |
- |
- |
| 7 |
Hb_025457_010 |
0.3118025129 |
- |
- |
- |
| 8 |
Hb_001703_120 |
0.3130588501 |
- |
- |
PREDICTED: uncharacterized protein LOC105116966 isoform X2 [Populus euphratica] |
| 9 |
Hb_000165_150 |
0.3182766586 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 10 |
Hb_176825_010 |
0.3190895663 |
- |
- |
PREDICTED: uncharacterized protein LOC105630857 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005656_060 |
0.325724674 |
- |
- |
- |
| 12 |
Hb_123521_010 |
0.3287442487 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 13 |
Hb_002686_380 |
0.3342230758 |
- |
- |
- |
| 14 |
Hb_140572_010 |
0.3398433172 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 15 |
Hb_038011_010 |
0.3408967635 |
- |
- |
hypothetical protein POPTR_0031s004401g, partial [Populus trichocarpa] |
| 16 |
Hb_001650_010 |
0.3419753366 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 17 |
Hb_010317_020 |
0.3437082142 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 18 |
Hb_000422_120 |
0.3440962514 |
- |
- |
PREDICTED: 1-phosphatidylinositol-3-phosphate 5-kinase FAB1B [Jatropha curcas] |
| 19 |
Hb_015707_010 |
0.3466178824 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 20 |
Hb_001545_210 |
0.3468943062 |
- |
- |
hypothetical protein JCGZ_11132 [Jatropha curcas] |