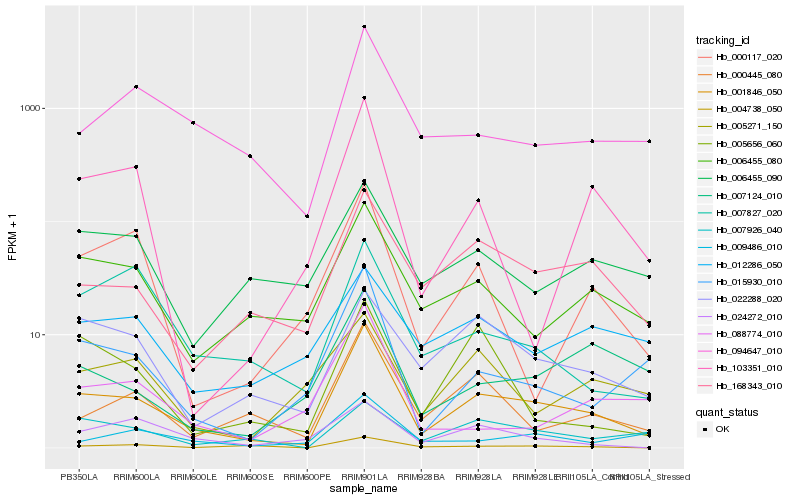
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001846_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_168343_010 |
0.2094563476 |
- |
- |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 3 |
Hb_015930_010 |
0.2099890369 |
- |
- |
uncharacterized protein LOC100499930 [Glycine max] |
| 4 |
Hb_000445_080 |
0.2120956768 |
transcription factor |
TF Family: MIKC |
PREDICTED: floral homeotic protein PMADS 2 [Jatropha curcas] |
| 5 |
Hb_006455_080 |
0.2204010591 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 6 |
Hb_094647_010 |
0.2204075039 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B-like [Jatropha curcas] |
| 7 |
Hb_004738_050 |
0.2299378471 |
- |
- |
F-box/leucine rich repeat protein, putative [Ricinus communis] |
| 8 |
Hb_103351_010 |
0.2352772104 |
- |
- |
hypothetical protein POPTR_0002s01570g [Populus trichocarpa] |
| 9 |
Hb_007124_010 |
0.241409997 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_022288_020 |
0.24272821 |
- |
- |
hypothetical protein POPTR_0006s08590g [Populus trichocarpa] |
| 11 |
Hb_009486_010 |
0.2431932308 |
- |
- |
- |
| 12 |
Hb_000117_020 |
0.2443132455 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 13 |
Hb_088774_010 |
0.247939512 |
- |
- |
- |
| 14 |
Hb_007827_020 |
0.2480873626 |
- |
- |
- |
| 15 |
Hb_006455_090 |
0.2506528892 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 16 |
Hb_007926_040 |
0.2537289034 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 17 |
Hb_024272_010 |
0.2564673917 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 18 |
Hb_005271_150 |
0.2609218458 |
- |
- |
PREDICTED: EKC/KEOPS complex subunit Tprkb-like [Malus domestica] |
| 19 |
Hb_005656_060 |
0.2623368649 |
- |
- |
- |
| 20 |
Hb_012286_050 |
0.2623599612 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |