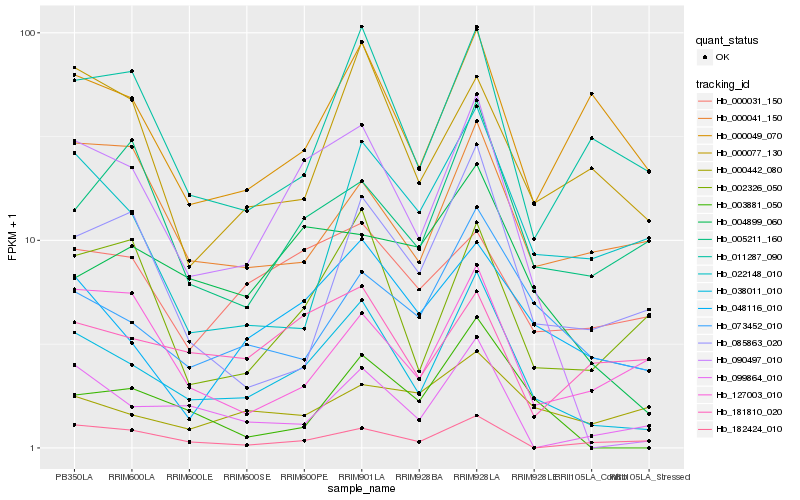
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038011_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0031s004401g, partial [Populus trichocarpa] |
| 2 |
Hb_090497_010 |
0.1515869668 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 3 |
Hb_073452_010 |
0.1776626363 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 4 |
Hb_048116_010 |
0.1853297839 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 5 |
Hb_011287_090 |
0.1928031304 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 6 |
Hb_099864_010 |
0.1951176284 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 7 |
Hb_002326_050 |
0.2029540471 |
- |
- |
PREDICTED: uncharacterized protein LOC105633701 [Jatropha curcas] |
| 8 |
Hb_004899_060 |
0.2037653408 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 9 |
Hb_003881_050 |
0.2048498991 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 10 |
Hb_085863_020 |
0.2073261385 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 11 |
Hb_000077_130 |
0.2109593501 |
- |
- |
PREDICTED: callose synthase 12 [Jatropha curcas] |
| 12 |
Hb_000049_070 |
0.2124399723 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |
| 13 |
Hb_022148_010 |
0.2140788044 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 14 |
Hb_005211_160 |
0.2206707149 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 15 |
Hb_182424_010 |
0.2232794814 |
- |
- |
PREDICTED: U-box domain-containing protein 51 [Jatropha curcas] |
| 16 |
Hb_000031_150 |
0.226040294 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 17 |
Hb_127003_010 |
0.2280026122 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 18 |
Hb_000041_150 |
0.229160795 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 19 |
Hb_181810_020 |
0.2322649078 |
- |
- |
- |
| 20 |
Hb_000442_080 |
0.2337619886 |
- |
- |
hypothetical protein JCGZ_03373 [Jatropha curcas] |