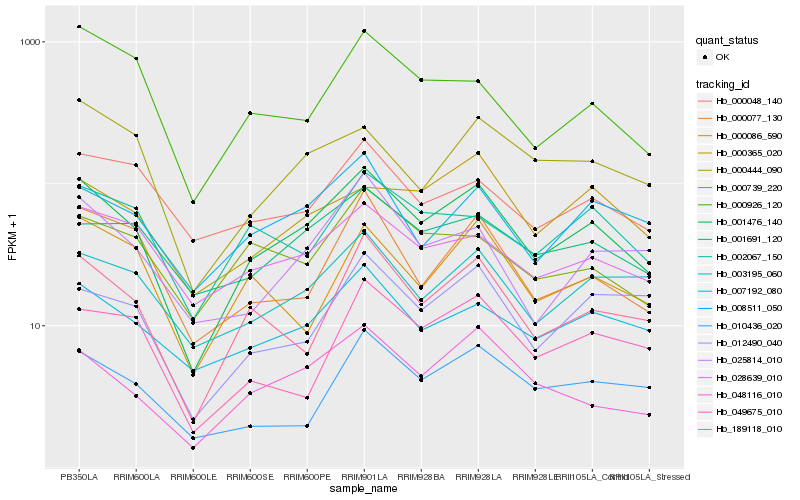
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001476_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630497 [Jatropha curcas] |
| 2 |
Hb_048116_010 |
0.1194917464 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 3 |
Hb_189118_010 |
0.1210534857 |
- |
- |
PREDICTED: extended synaptotagmin-1 [Cucumis melo] |
| 4 |
Hb_007192_080 |
0.1254768559 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_008511_050 |
0.1315733472 |
- |
- |
PREDICTED: cyclase-associated protein 1 [Jatropha curcas] |
| 6 |
Hb_028639_010 |
0.1335253907 |
- |
- |
transporter, putative [Ricinus communis] |
| 7 |
Hb_003195_060 |
0.1356583262 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 8 |
Hb_000077_130 |
0.1383311692 |
- |
- |
PREDICTED: callose synthase 12 [Jatropha curcas] |
| 9 |
Hb_001691_120 |
0.1383767039 |
desease resistance |
Gene Name: AAA |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |
| 10 |
Hb_002067_150 |
0.1395647306 |
- |
- |
PREDICTED: ankyrin repeat protein SKIP35 [Jatropha curcas] |
| 11 |
Hb_000444_090 |
0.1401905789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_025814_010 |
0.1414048053 |
- |
- |
hypothetical protein PRUPE_ppa015585mg [Prunus persica] |
| 13 |
Hb_000365_020 |
0.1441004936 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000739_220 |
0.145572765 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 15 |
Hb_000086_590 |
0.1471214385 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 16 |
Hb_049675_010 |
0.1472265261 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 17 |
Hb_010436_020 |
0.1473366677 |
- |
- |
Dicer-like protein 4 [Morus notabilis] |
| 18 |
Hb_000926_120 |
0.1473796533 |
- |
- |
PREDICTED: aspartic proteinase A1-like [Jatropha curcas] |
| 19 |
Hb_012490_040 |
0.1478406537 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 20 |
Hb_000048_140 |
0.1479373209 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |