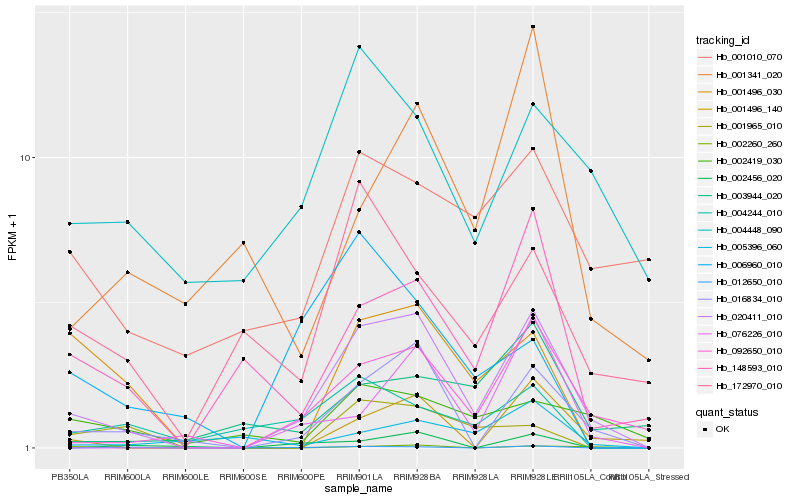
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020411_010 |
0.0 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 2 |
Hb_016834_010 |
0.2041275612 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 3 |
Hb_092650_010 |
0.2148494018 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 4 |
Hb_148593_010 |
0.2764143396 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR-like [Jatropha curcas] |
| 5 |
Hb_001496_140 |
0.2920907179 |
- |
- |
- |
| 6 |
Hb_076226_010 |
0.2958124017 |
- |
- |
hypothetical protein VITISV_030432 [Vitis vinifera] |
| 7 |
Hb_172970_010 |
0.3046815173 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 8 |
Hb_002419_030 |
0.3063809008 |
- |
- |
- |
| 9 |
Hb_001496_030 |
0.3095210368 |
- |
- |
hypothetical protein CISIN_1g0170981mg, partial [Citrus sinensis] |
| 10 |
Hb_004448_090 |
0.3096479627 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 11 |
Hb_004244_010 |
0.3169174072 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 12 |
Hb_003944_020 |
0.3183746235 |
- |
- |
PREDICTED: uncharacterized protein LOC105110580 [Populus euphratica] |
| 13 |
Hb_012650_010 |
0.3193233376 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 14 |
Hb_001010_070 |
0.3199189116 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 15 |
Hb_001341_020 |
0.3229939097 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 16 |
Hb_005396_060 |
0.3252464622 |
- |
- |
- |
| 17 |
Hb_006960_010 |
0.3261053445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001965_010 |
0.3270407956 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 19 |
Hb_002456_020 |
0.3281163901 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 20 |
Hb_002260_260 |
0.3286902698 |
- |
- |
hypothetical protein VITISV_037667 [Vitis vinifera] |