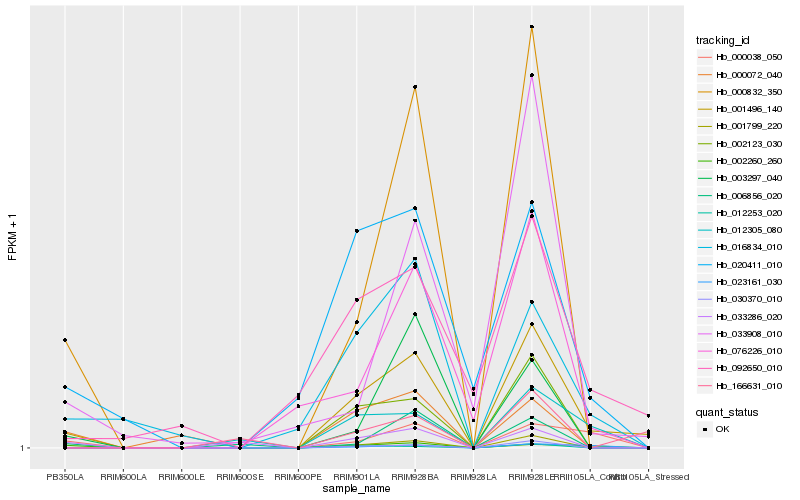
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001496_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_000832_350 |
0.2410953923 |
- |
- |
Thylakoid lumenal 25.6 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_001799_220 |
0.2590014551 |
transcription factor |
TF Family: AP2 |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 4 |
Hb_002123_030 |
0.2642406559 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 5 |
Hb_016834_010 |
0.2762962197 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 6 |
Hb_030370_010 |
0.2768429044 |
- |
- |
PREDICTED: uncharacterized protein LOC104896222 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_000038_050 |
0.27891602 |
- |
- |
- |
| 8 |
Hb_092650_010 |
0.2845658891 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 9 |
Hb_012253_020 |
0.2883647187 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_020411_010 |
0.2920907179 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_002260_260 |
0.2936950829 |
- |
- |
hypothetical protein VITISV_037667 [Vitis vinifera] |
| 12 |
Hb_012305_080 |
0.2939054156 |
- |
- |
- |
| 13 |
Hb_003297_040 |
0.3013830709 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 14 |
Hb_023161_030 |
0.3042298493 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 15 |
Hb_000072_040 |
0.3078587098 |
- |
- |
- |
| 16 |
Hb_076226_010 |
0.3079848856 |
- |
- |
hypothetical protein VITISV_030432 [Vitis vinifera] |
| 17 |
Hb_033908_010 |
0.3085468833 |
- |
- |
- |
| 18 |
Hb_166631_010 |
0.3117790951 |
- |
- |
PREDICTED: uncharacterized protein LOC105801076 [Gossypium raimondii] |
| 19 |
Hb_033286_020 |
0.3128630523 |
- |
- |
- |
| 20 |
Hb_006856_020 |
0.3161278599 |
- |
- |
- |