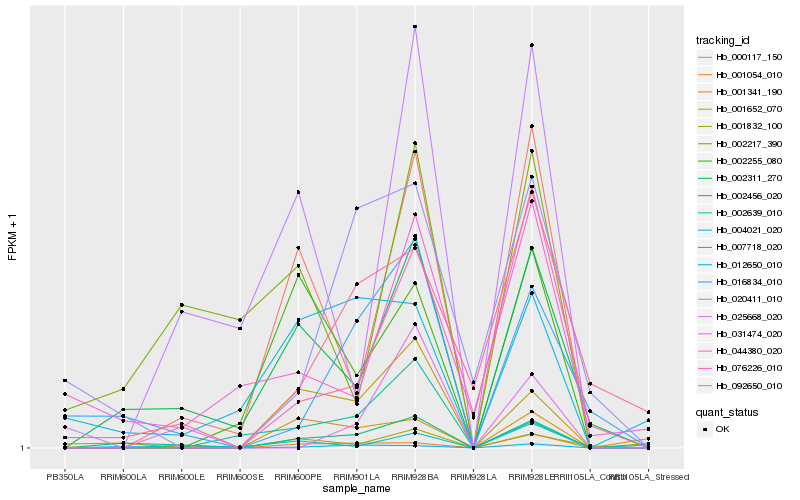
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002456_020 |
0.0 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 2 |
Hb_002311_270 |
0.1984244619 |
- |
- |
PREDICTED: WD repeat-containing protein 43-like [Malus domestica] |
| 3 |
Hb_012650_010 |
0.235291631 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 4 |
Hb_001652_070 |
0.2574659739 |
- |
- |
PREDICTED: uncharacterized protein LOC103439285 [Malus domestica] |
| 5 |
Hb_016834_010 |
0.2581511392 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 6 |
Hb_000117_150 |
0.264046809 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 7 |
Hb_001832_100 |
0.2721923492 |
- |
- |
- |
| 8 |
Hb_007718_020 |
0.3107281976 |
- |
- |
PREDICTED: receptor-like protein 12 [Camelina sativa] |
| 9 |
Hb_002255_080 |
0.3113213721 |
- |
- |
- |
| 10 |
Hb_076226_010 |
0.3144810634 |
- |
- |
hypothetical protein VITISV_030432 [Vitis vinifera] |
| 11 |
Hb_031474_020 |
0.3214111542 |
- |
- |
- |
| 12 |
Hb_025668_020 |
0.3223962063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_092650_010 |
0.3232608738 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 14 |
Hb_002217_390 |
0.327414832 |
- |
- |
PREDICTED: galactokinase [Jatropha curcas] |
| 15 |
Hb_020411_010 |
0.3281163901 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 16 |
Hb_001341_190 |
0.3288855491 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_004021_020 |
0.3301160758 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 18 |
Hb_002639_010 |
0.3324021387 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 19 |
Hb_044380_020 |
0.3328388253 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 20 |
Hb_001054_010 |
0.3338871498 |
- |
- |
hypothetical protein VITISV_011091 [Vitis vinifera] |