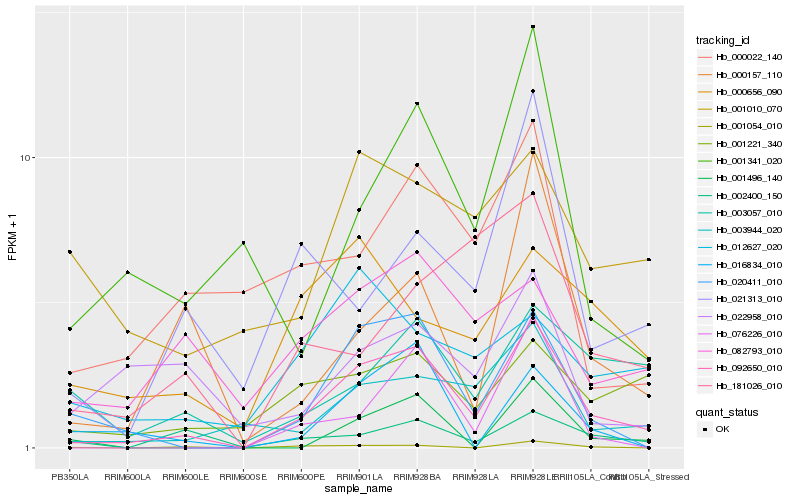
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092650_010 |
0.0 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 2 |
Hb_020411_010 |
0.2148494018 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 3 |
Hb_003944_020 |
0.2233746757 |
- |
- |
PREDICTED: uncharacterized protein LOC105110580 [Populus euphratica] |
| 4 |
Hb_001221_340 |
0.2568700288 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 5 |
Hb_000157_110 |
0.259720536 |
- |
- |
hypothetical protein Csa_6G439410 [Cucumis sativus] |
| 6 |
Hb_002400_150 |
0.2607232987 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 7 |
Hb_076226_010 |
0.2631229643 |
- |
- |
hypothetical protein VITISV_030432 [Vitis vinifera] |
| 8 |
Hb_082793_010 |
0.2658805165 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_021313_010 |
0.2699267025 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 10 |
Hb_003057_010 |
0.2716631282 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 11 |
Hb_001341_020 |
0.2717036688 |
- |
- |
Uncharacterised protein [Bordetella pertussis] |
| 12 |
Hb_000022_140 |
0.2750167012 |
- |
- |
hypothetical protein MIMGU_mgv1a009920mg [Erythranthe guttata] |
| 13 |
Hb_000656_090 |
0.2760401051 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 14 |
Hb_016834_010 |
0.2775935573 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 15 |
Hb_001054_010 |
0.2790446088 |
- |
- |
hypothetical protein VITISV_011091 [Vitis vinifera] |
| 16 |
Hb_012627_020 |
0.2810605051 |
- |
- |
PHYTOCHROME AND FLOWERING TIME 1 family protein [Populus trichocarpa] |
| 17 |
Hb_001496_140 |
0.2845658891 |
- |
- |
- |
| 18 |
Hb_001010_070 |
0.2855244693 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 19 |
Hb_181026_010 |
0.2883909512 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 20 |
Hb_022958_010 |
0.2901666109 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Populus euphratica] |