| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
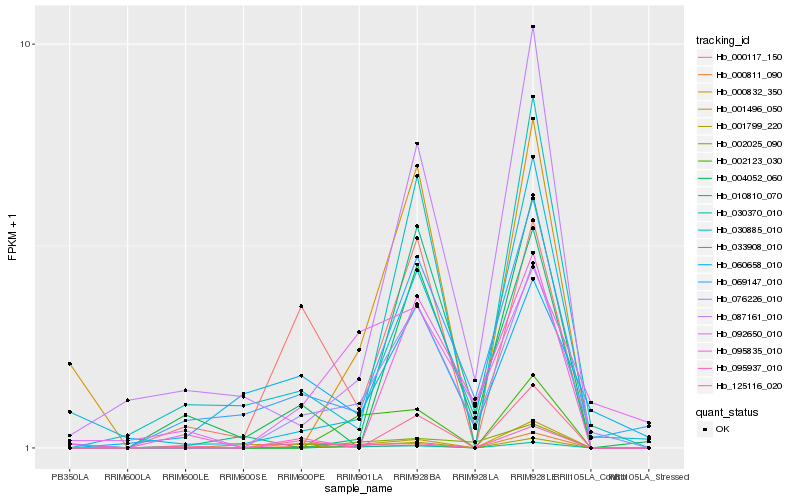
Hb_076226_010 |
0.0 |
- |
- |
hypothetical protein VITISV_030432 [Vitis vinifera] |
| 2 |
Hb_069147_010 |
0.2250844889 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 3 |
Hb_033908_010 |
0.2381741605 |
- |
- |
- |
| 4 |
Hb_001799_220 |
0.250175281 |
transcription factor |
TF Family: AP2 |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 5 |
Hb_092650_010 |
0.2631229643 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 6 |
Hb_030885_010 |
0.2645255219 |
- |
- |
- |
| 7 |
Hb_087161_010 |
0.2687263771 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_002025_090 |
0.2729706314 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_010810_070 |
0.276509275 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 10 |
Hb_030370_010 |
0.2784222852 |
- |
- |
PREDICTED: uncharacterized protein LOC104896222 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_095835_010 |
0.2791580421 |
- |
- |
hypothetical protein JCGZ_23410 [Jatropha curcas] |
| 12 |
Hb_000117_150 |
0.281954683 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 13 |
Hb_004052_060 |
0.2821256794 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_060658_010 |
0.282537318 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_125116_020 |
0.2849763496 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X5 [Populus euphratica] |
| 16 |
Hb_000811_090 |
0.2864499219 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 17 |
Hb_002123_030 |
0.2906570974 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 18 |
Hb_000832_350 |
0.2913175666 |
- |
- |
Thylakoid lumenal 25.6 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_095937_010 |
0.2915109411 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 20 |
Hb_001496_050 |
0.2922977661 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |