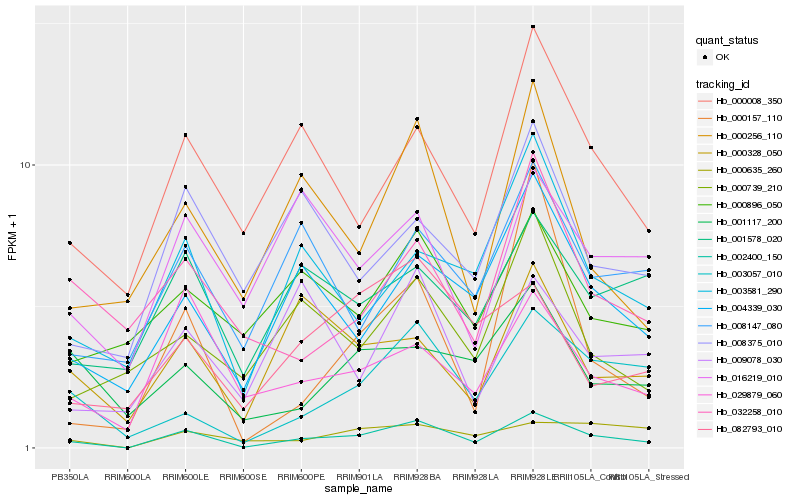
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004339_030 |
0.2029497606 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 3 |
Hb_003581_290 |
0.2106020485 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 3 [Fragaria vesca subsp. vesca] |
| 4 |
Hb_000256_110 |
0.2126061381 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003057_010 |
0.2141475869 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 6 |
Hb_029879_060 |
0.2142276918 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 7 |
Hb_000008_350 |
0.2160304266 |
- |
- |
- |
| 8 |
Hb_000328_050 |
0.2255491364 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000157_110 |
0.2282142132 |
- |
- |
hypothetical protein Csa_6G439410 [Cucumis sativus] |
| 10 |
Hb_000739_210 |
0.2341019111 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g01970 [Jatropha curcas] |
| 11 |
Hb_008147_080 |
0.2354925231 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001578_020 |
0.236432839 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 13 |
Hb_082793_010 |
0.2378701812 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000635_260 |
0.2393583118 |
- |
- |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 15 |
Hb_008375_010 |
0.2409186278 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 16 |
Hb_001117_200 |
0.2419075355 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000896_050 |
0.2422209442 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 18 |
Hb_032258_010 |
0.2427157814 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 19 |
Hb_016219_010 |
0.2427176945 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |
| 20 |
Hb_009078_030 |
0.2429861565 |
- |
- |
hypothetical protein PRUPE_ppa016992mg, partial [Prunus persica] |