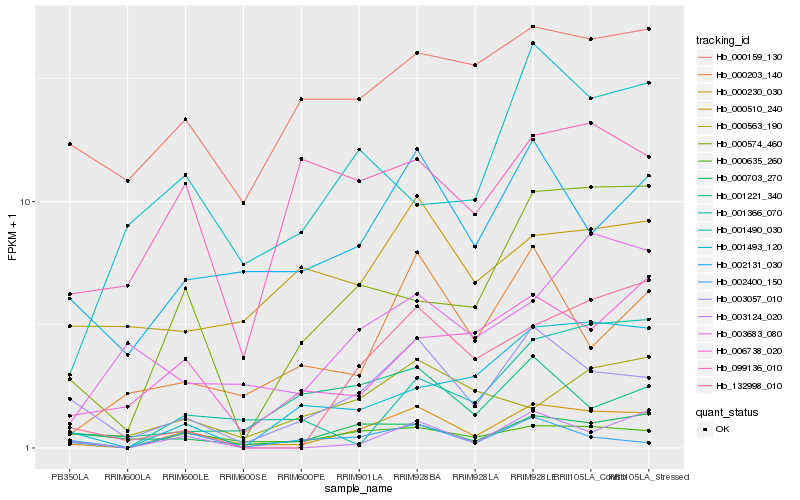
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_030 |
0.0 |
- |
- |
hypothetical protein B456_013G2021001, partial [Gossypium raimondii] |
| 2 |
Hb_000635_260 |
0.1793052308 |
- |
- |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 3 |
Hb_003057_010 |
0.1875174429 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 4 |
Hb_000574_460 |
0.2074630424 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 5 |
Hb_002131_030 |
0.2183669225 |
- |
- |
PREDICTED: pyruvate kinase isozyme G, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003124_020 |
0.2233971766 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 7 |
Hb_132998_010 |
0.2240021872 |
- |
- |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 8 |
Hb_006738_020 |
0.2363758946 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 9 |
Hb_000203_140 |
0.2375588483 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 10 |
Hb_000703_270 |
0.2417732519 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 11 |
Hb_001493_120 |
0.2418393629 |
- |
- |
- |
| 12 |
Hb_000563_190 |
0.2439784921 |
- |
- |
hypothetical protein PRUPE_ppa002828mg [Prunus persica] |
| 13 |
Hb_000159_130 |
0.2547627537 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_001490_030 |
0.257173163 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003683_080 |
0.2610066286 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 16 |
Hb_001366_070 |
0.2610803112 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 4-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_099136_010 |
0.2615669227 |
- |
- |
PREDICTED: cysteine synthase-like isoform X3 [Cucumis sativus] |
| 18 |
Hb_002400_150 |
0.2631491284 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001221_340 |
0.266120042 |
- |
- |
hypothetical protein EUGRSUZ_B00102 [Eucalyptus grandis] |
| 20 |
Hb_000510_240 |
0.267347802 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |