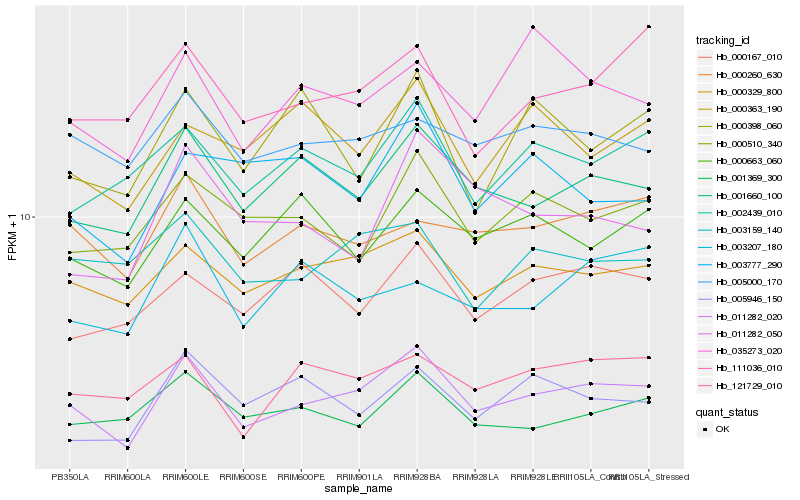
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011282_020 |
0.0 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001660_100 |
0.1140991633 |
- |
- |
PREDICTED: glyoxylate reductase/hydroxypyruvate reductase [Jatropha curcas] |
| 3 |
Hb_000329_800 |
0.1198313435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_121729_010 |
0.1321397426 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000260_630 |
0.1324212794 |
- |
- |
PREDICTED: altered inheritance rate of mitochondria protein 25 isoform X1 [Jatropha curcas] |
| 6 |
Hb_111036_010 |
0.1333112102 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 7 |
Hb_002439_010 |
0.1343481487 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 8 |
Hb_000510_340 |
0.1357738871 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 9 |
Hb_001369_300 |
0.1362717964 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 10 |
Hb_011282_050 |
0.1363495544 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_035273_020 |
0.1369505475 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000663_060 |
0.1396149394 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 13 |
Hb_000167_010 |
0.1401880348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005000_170 |
0.1422295036 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 15 |
Hb_003159_140 |
0.1423605653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000363_190 |
0.1441350063 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003207_180 |
0.1443939649 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000398_060 |
0.1460057025 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 19 |
Hb_005946_150 |
0.1461075362 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 20 |
Hb_003777_290 |
0.1463932501 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |