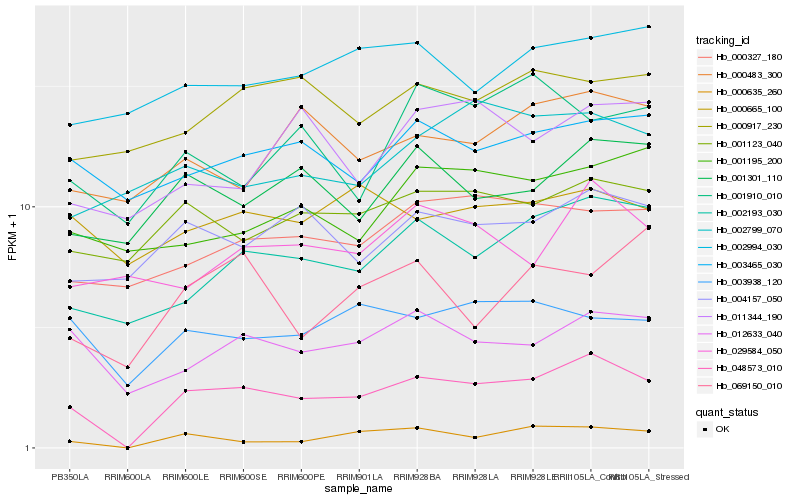
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048573_010 |
0.0 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 2 |
Hb_000635_260 |
0.1479036472 |
- |
- |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 3 |
Hb_002193_030 |
0.1622487573 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 4 |
Hb_012633_040 |
0.1727521202 |
- |
- |
- |
| 5 |
Hb_001123_040 |
0.1799555312 |
- |
- |
PREDICTED: R3H domain-containing protein 1-like [Jatropha curcas] |
| 6 |
Hb_000327_180 |
0.1818815877 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004157_050 |
0.182726541 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001301_110 |
0.1830003218 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 9 |
Hb_003938_120 |
0.1845462662 |
- |
- |
autophagy 9 family protein, partial [Populus trichocarpa] |
| 10 |
Hb_003465_030 |
0.1884893626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000483_300 |
0.1908302742 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 12 |
Hb_001195_200 |
0.195829126 |
- |
- |
PREDICTED: uncharacterized protein LOC105633802 [Jatropha curcas] |
| 13 |
Hb_000917_230 |
0.1962507224 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Jatropha curcas] |
| 14 |
Hb_029584_050 |
0.196412548 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3 [Jatropha curcas] |
| 15 |
Hb_011344_190 |
0.1967549955 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 16 |
Hb_001910_010 |
0.1968055376 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_069150_010 |
0.1970048952 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 18 |
Hb_002994_030 |
0.1978410339 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-like [Pyrus x bretschneideri] |
| 19 |
Hb_002799_070 |
0.1981729161 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 20 |
Hb_000665_100 |
0.1984221329 |
- |
- |
PREDICTED: neurochondrin [Jatropha curcas] |