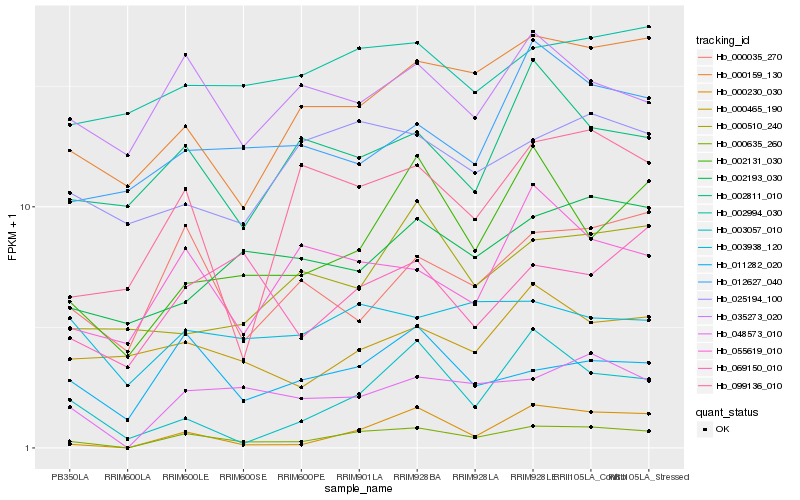
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 2 |
Hb_048573_010 |
0.1479036472 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 3 |
Hb_000230_030 |
0.1793052308 |
- |
- |
hypothetical protein B456_013G2021001, partial [Gossypium raimondii] |
| 4 |
Hb_000159_130 |
0.1822880483 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_002131_030 |
0.1888580434 |
- |
- |
PREDICTED: pyruvate kinase isozyme G, chloroplastic [Jatropha curcas] |
| 6 |
Hb_011282_020 |
0.190847231 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_099136_010 |
0.1984010384 |
- |
- |
PREDICTED: cysteine synthase-like isoform X3 [Cucumis sativus] |
| 8 |
Hb_000035_270 |
0.2004363393 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |
| 9 |
Hb_055619_010 |
0.2027199718 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002193_030 |
0.2072582292 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 11 |
Hb_025194_100 |
0.2108858428 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 12 |
Hb_002811_010 |
0.2125388927 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 13 |
Hb_002994_030 |
0.2130275714 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-like [Pyrus x bretschneideri] |
| 14 |
Hb_035273_020 |
0.2135839093 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003057_010 |
0.2146896731 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 16 |
Hb_012627_040 |
0.2148672489 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 17 |
Hb_003938_120 |
0.2152361985 |
- |
- |
autophagy 9 family protein, partial [Populus trichocarpa] |
| 18 |
Hb_000510_240 |
0.2154473825 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_069150_010 |
0.2155714596 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 20 |
Hb_000465_190 |
0.2156394402 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X2 [Jatropha curcas] |