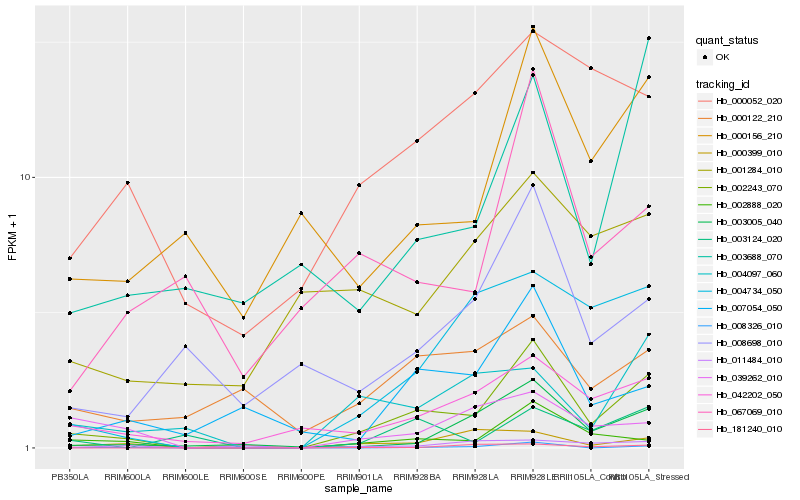
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002243_070 |
0.0 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 2 |
Hb_042202_050 |
0.2451449231 |
- |
- |
TPA: hypothetical protein ZEAMMB73_683745 [Zea mays] |
| 3 |
Hb_002888_020 |
0.2455136927 |
- |
- |
- |
| 4 |
Hb_004734_050 |
0.2465800984 |
- |
- |
- |
| 5 |
Hb_003005_040 |
0.2560047391 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_004097_060 |
0.2820857077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000156_210 |
0.2857959505 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 8 |
Hb_039262_010 |
0.2890647343 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 9 |
Hb_003688_070 |
0.2897273021 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 10 |
Hb_003124_020 |
0.2911292657 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 11 |
Hb_007054_050 |
0.2923486369 |
- |
- |
- |
| 12 |
Hb_000052_020 |
0.2943248201 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 13 |
Hb_008698_010 |
0.3034416553 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_067069_010 |
0.3042715034 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 15 |
Hb_181240_010 |
0.3054489605 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 16 |
Hb_000399_010 |
0.3097844877 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 17 |
Hb_008326_010 |
0.3098685579 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 18 |
Hb_000122_210 |
0.3106591722 |
- |
- |
- |
| 19 |
Hb_001284_010 |
0.3137825281 |
- |
- |
hypothetical protein CISIN_1g0279981mg, partial [Citrus sinensis] |
| 20 |
Hb_011484_010 |
0.313914592 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |