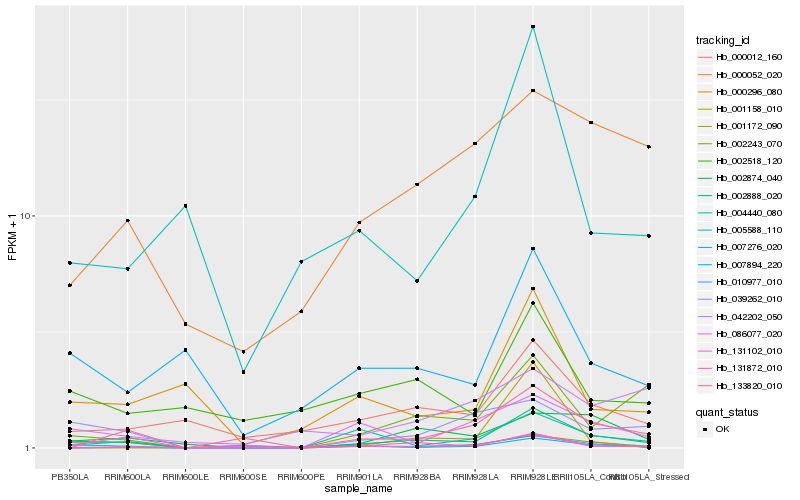
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002888_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002243_070 |
0.2455136927 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 3 |
Hb_039262_010 |
0.2469811773 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_002874_040 |
0.2474471686 |
- |
- |
- |
| 5 |
Hb_004440_080 |
0.2607260683 |
- |
- |
PREDICTED: syntaxin-112 [Populus euphratica] |
| 6 |
Hb_086077_020 |
0.2721738481 |
- |
- |
hypothetical protein PHAVU_001G054900g [Phaseolus vulgaris] |
| 7 |
Hb_000012_160 |
0.2745369265 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 8 |
Hb_133820_010 |
0.2800335996 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 9 |
Hb_131102_010 |
0.2806391035 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_131872_010 |
0.2868007498 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_001172_090 |
0.2883194791 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 12 |
Hb_007276_020 |
0.2887655096 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 13 |
Hb_007894_220 |
0.2910707697 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 14 |
Hb_000052_020 |
0.2936109227 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 15 |
Hb_010977_010 |
0.2966407349 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_000296_080 |
0.2970144995 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35130 [Jatropha curcas] |
| 17 |
Hb_005588_110 |
0.2988440004 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 18 |
Hb_001158_010 |
0.2991671128 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 19 |
Hb_042202_050 |
0.3044776167 |
- |
- |
TPA: hypothetical protein ZEAMMB73_683745 [Zea mays] |
| 20 |
Hb_002518_120 |
0.3057799989 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |