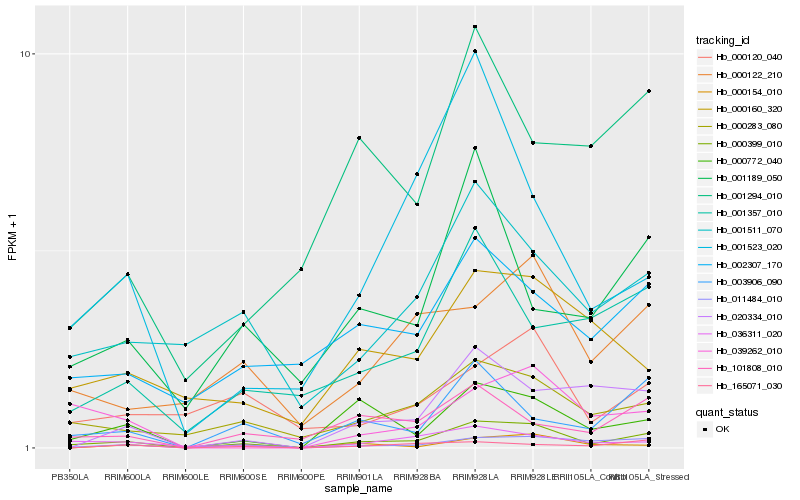
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000399_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 2 |
Hb_011484_010 |
0.1848811578 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 3 |
Hb_000772_040 |
0.1962057578 |
- |
- |
PREDICTED: uncharacterized protein LOC105036273 [Elaeis guineensis] |
| 4 |
Hb_000283_080 |
0.2169379917 |
- |
- |
- |
| 5 |
Hb_036311_020 |
0.2248838554 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_003906_090 |
0.229129382 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 7 |
Hb_001523_020 |
0.2372119275 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |
| 8 |
Hb_101808_010 |
0.2440503159 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 9 |
Hb_020334_010 |
0.2487331726 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 10 |
Hb_000122_210 |
0.2500939201 |
- |
- |
- |
| 11 |
Hb_001511_070 |
0.2522441898 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 12 |
Hb_001357_010 |
0.2531244265 |
- |
- |
- |
| 13 |
Hb_001189_050 |
0.2572308816 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 14 |
Hb_039262_010 |
0.2605261157 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_165071_030 |
0.2620343139 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_000154_010 |
0.2625121758 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_000160_320 |
0.2633646274 |
- |
- |
hypothetical protein POPTR_0004s08730g [Populus trichocarpa] |
| 18 |
Hb_002307_170 |
0.264180647 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 19 |
Hb_000120_040 |
0.2644618269 |
- |
- |
- |
| 20 |
Hb_001294_010 |
0.2689407187 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |