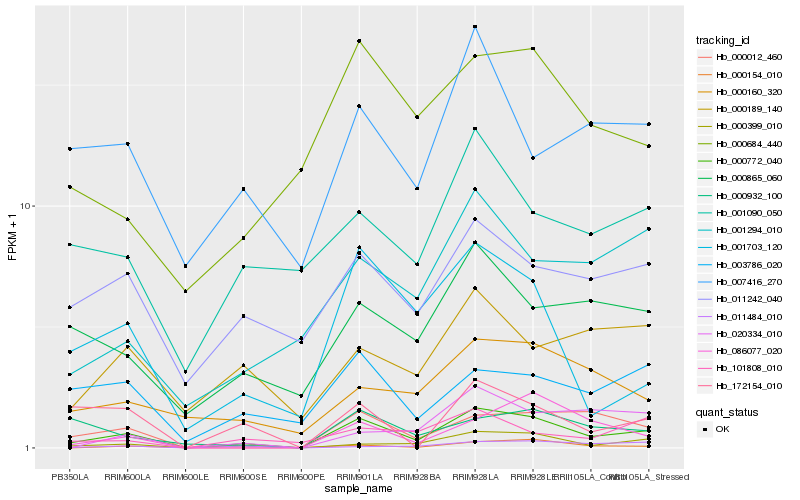
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000772_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105036273 [Elaeis guineensis] |
| 2 |
Hb_000399_010 |
0.1962057578 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 3 |
Hb_000012_460 |
0.2122744295 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_001703_120 |
0.2344909207 |
- |
- |
PREDICTED: uncharacterized protein LOC105116966 isoform X2 [Populus euphratica] |
| 5 |
Hb_000865_060 |
0.2424219003 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 6 |
Hb_001294_010 |
0.2436563047 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 7 |
Hb_000160_320 |
0.2479446121 |
- |
- |
hypothetical protein POPTR_0004s08730g [Populus trichocarpa] |
| 8 |
Hb_003786_020 |
0.2498159428 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 9 |
Hb_086077_020 |
0.2513131142 |
- |
- |
hypothetical protein PHAVU_001G054900g [Phaseolus vulgaris] |
| 10 |
Hb_000932_100 |
0.2514732353 |
- |
- |
- |
| 11 |
Hb_000684_440 |
0.25234947 |
- |
- |
hypothetical protein CICLE_v100111082mg, partial [Citrus clementina] |
| 12 |
Hb_101808_010 |
0.2533414233 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000154_010 |
0.2533488336 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_011242_040 |
0.2536506666 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007416_270 |
0.2571648617 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 16 |
Hb_172154_010 |
0.2578756648 |
- |
- |
hypothetical protein JCGZ_25247 [Jatropha curcas] |
| 17 |
Hb_000189_140 |
0.2595795376 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_020334_010 |
0.259937353 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 19 |
Hb_011484_010 |
0.2614431577 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 20 |
Hb_001090_050 |
0.2632650473 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |