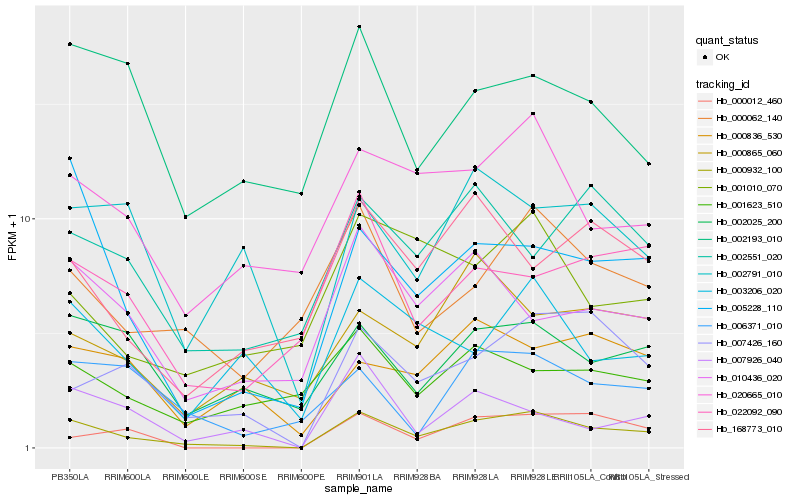
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000932_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_005228_110 |
0.1987658436 |
- |
- |
Cleavage and polyadenylation specificity factor subunit 3 [Fukomys damarensis] |
| 3 |
Hb_007426_160 |
0.200155654 |
- |
- |
- |
| 4 |
Hb_020665_010 |
0.2077454653 |
- |
- |
PREDICTED: uncharacterized protein LOC103438977 isoform X2 [Malus domestica] |
| 5 |
Hb_000062_140 |
0.2082263491 |
transcription factor |
TF Family: GRAS |
hypothetical protein JCGZ_17897 [Jatropha curcas] |
| 6 |
Hb_002025_200 |
0.2092864069 |
- |
- |
- |
| 7 |
Hb_003206_020 |
0.2122969577 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 8 |
Hb_010436_020 |
0.2139811647 |
- |
- |
Dicer-like protein 4 [Morus notabilis] |
| 9 |
Hb_022092_090 |
0.2202822571 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 10 |
Hb_000012_460 |
0.2211958159 |
- |
- |
unknown [Populus trichocarpa] |
| 11 |
Hb_002791_010 |
0.2259913739 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_007926_040 |
0.2261089133 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 13 |
Hb_168773_010 |
0.2274179911 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_006371_010 |
0.2280905049 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 15 |
Hb_000836_530 |
0.2281312566 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 16 |
Hb_001010_070 |
0.2297958716 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 17 |
Hb_002193_010 |
0.2301796794 |
- |
- |
PREDICTED: SUN domain-containing protein 3-like [Jatropha curcas] |
| 18 |
Hb_001623_510 |
0.2301813284 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_002551_020 |
0.2304201317 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000865_060 |
0.2319626802 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |