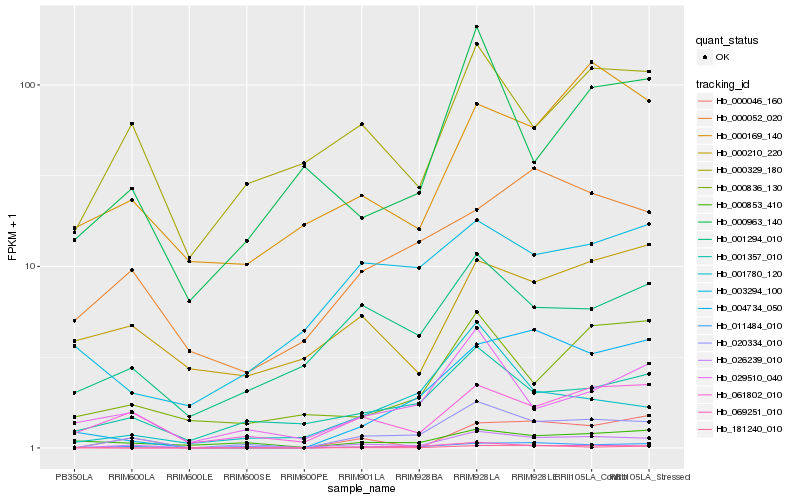
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026239_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_020334_010 |
0.0979492557 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_011484_010 |
0.1967799714 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 4 |
Hb_061802_010 |
0.2091381327 |
- |
- |
hypothetical protein VITISV_011986 [Vitis vinifera] |
| 5 |
Hb_001357_010 |
0.2219637849 |
- |
- |
- |
| 6 |
Hb_000046_160 |
0.2254363422 |
- |
- |
Pentatricopeptide repeat-containing protein, mitochondrial [Glycine soja] |
| 7 |
Hb_001294_010 |
0.23117808 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 8 |
Hb_000329_180 |
0.2358915545 |
- |
- |
Auxin-induced protein X10A, putative [Ricinus communis] |
| 9 |
Hb_000169_140 |
0.2383008481 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 10 |
Hb_000836_130 |
0.24049686 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004734_050 |
0.2412908867 |
- |
- |
- |
| 12 |
Hb_000963_140 |
0.2453179468 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000052_020 |
0.246725693 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 14 |
Hb_029510_040 |
0.2491923174 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 15 |
Hb_069251_010 |
0.2516632948 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_003294_100 |
0.258726798 |
- |
- |
hypothetical protein VITISV_009629 [Vitis vinifera] |
| 17 |
Hb_001780_120 |
0.2595678487 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 18 |
Hb_000210_220 |
0.262616826 |
- |
- |
hypothetical protein POPTR_0006s22010g [Populus trichocarpa] |
| 19 |
Hb_181240_010 |
0.2628805833 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 20 |
Hb_000853_410 |
0.263262825 |
- |
- |
- |