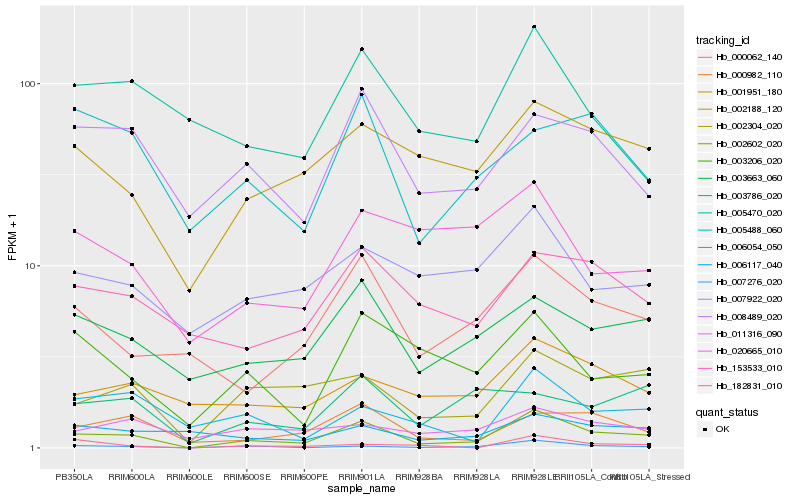
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002602_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_007276_020 |
0.1781514642 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 3 |
Hb_002304_020 |
0.2063155857 |
- |
- |
- |
| 4 |
Hb_000982_110 |
0.2104455843 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000062_140 |
0.2180093779 |
transcription factor |
TF Family: GRAS |
hypothetical protein JCGZ_17897 [Jatropha curcas] |
| 6 |
Hb_003663_060 |
0.2194514392 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g43820 [Jatropha curcas] |
| 7 |
Hb_002188_120 |
0.2225705235 |
- |
- |
PREDICTED: thymocyte nuclear protein 1 [Pyrus x bretschneideri] |
| 8 |
Hb_003206_020 |
0.2292161575 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 9 |
Hb_008489_020 |
0.2300541072 |
- |
- |
PREDICTED: uncharacterized protein LOC105644663 [Jatropha curcas] |
| 10 |
Hb_153533_010 |
0.2331204068 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X3 [Jatropha curcas] |
| 11 |
Hb_001951_180 |
0.2346246549 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_006054_050 |
0.2367358213 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 13 |
Hb_007922_020 |
0.2399513614 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 14 |
Hb_005470_020 |
0.2427588404 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_005488_060 |
0.2432835114 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_011316_090 |
0.2447446165 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 17 |
Hb_006117_040 |
0.2472026927 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 18 |
Hb_003786_020 |
0.2482939653 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 19 |
Hb_020665_010 |
0.2492356893 |
- |
- |
PREDICTED: uncharacterized protein LOC103438977 isoform X2 [Malus domestica] |
| 20 |
Hb_182831_010 |
0.2503737094 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |