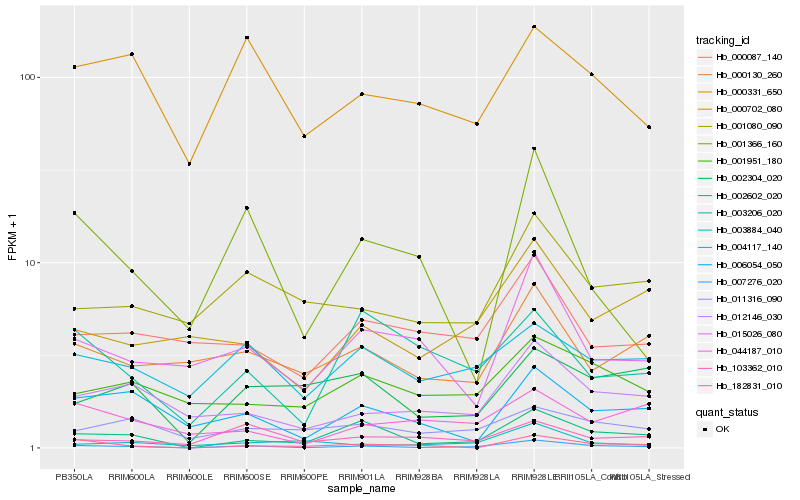
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007276_020 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_002602_020 |
0.1781514642 |
- |
- |
- |
| 3 |
Hb_012146_030 |
0.2312792384 |
- |
- |
- |
| 4 |
Hb_015026_080 |
0.2470923028 |
- |
- |
- |
| 5 |
Hb_182831_010 |
0.2522621752 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_006054_050 |
0.2527168511 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 7 |
Hb_000702_080 |
0.253211437 |
- |
- |
PREDICTED: uncharacterized protein LOC105630492 [Jatropha curcas] |
| 8 |
Hb_001366_160 |
0.2547092998 |
- |
- |
- |
| 9 |
Hb_103362_010 |
0.2606190772 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 10 |
Hb_001080_090 |
0.2620247978 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004117_140 |
0.2626854222 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g16710, mitochondrial [Populus euphratica] |
| 12 |
Hb_000331_650 |
0.2655824335 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase-like [Jatropha curcas] |
| 13 |
Hb_000087_140 |
0.2670783991 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001951_180 |
0.2676039357 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_003884_040 |
0.2678312018 |
- |
- |
putative gag-pol polyprotein [Oryza sativa Japonica Group] |
| 16 |
Hb_000130_260 |
0.2703766936 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002304_020 |
0.2714038395 |
- |
- |
- |
| 18 |
Hb_003206_020 |
0.2715533304 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 19 |
Hb_044187_010 |
0.274997269 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 20 |
Hb_011316_090 |
0.2753070684 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |