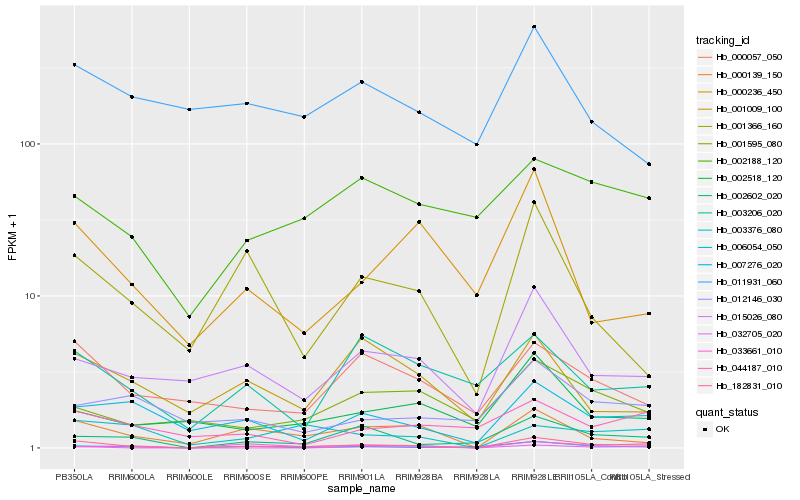
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182831_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_000139_150 |
0.233848243 |
- |
- |
transposon protein, putative, Mutator sub-class [Oryza sativa Japonica Group] |
| 3 |
Hb_002602_020 |
0.2503737094 |
- |
- |
- |
| 4 |
Hb_007276_020 |
0.2522621752 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_015026_080 |
0.2590979527 |
- |
- |
- |
| 6 |
Hb_006054_050 |
0.2594496005 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 7 |
Hb_000057_050 |
0.2600834558 |
- |
- |
hypothetical protein Csa_7G132920 [Cucumis sativus] |
| 8 |
Hb_001366_160 |
0.2699425369 |
- |
- |
- |
| 9 |
Hb_000236_450 |
0.2785971776 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 10 |
Hb_002518_120 |
0.2799601003 |
- |
- |
hypothetical protein RCOM_0731250 [Ricinus communis] |
| 11 |
Hb_003376_080 |
0.2859397399 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X1 [Jatropha curcas] |
| 12 |
Hb_032705_020 |
0.2891629221 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 13 |
Hb_002188_120 |
0.2893759903 |
- |
- |
PREDICTED: thymocyte nuclear protein 1 [Pyrus x bretschneideri] |
| 14 |
Hb_001595_080 |
0.2914788128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001009_100 |
0.2918643546 |
- |
- |
- |
| 16 |
Hb_033661_010 |
0.2923924249 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 17 |
Hb_003206_020 |
0.2952796532 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 18 |
Hb_012146_030 |
0.2956473554 |
- |
- |
- |
| 19 |
Hb_044187_010 |
0.2972506713 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 20 |
Hb_011931_060 |
0.2975811381 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |