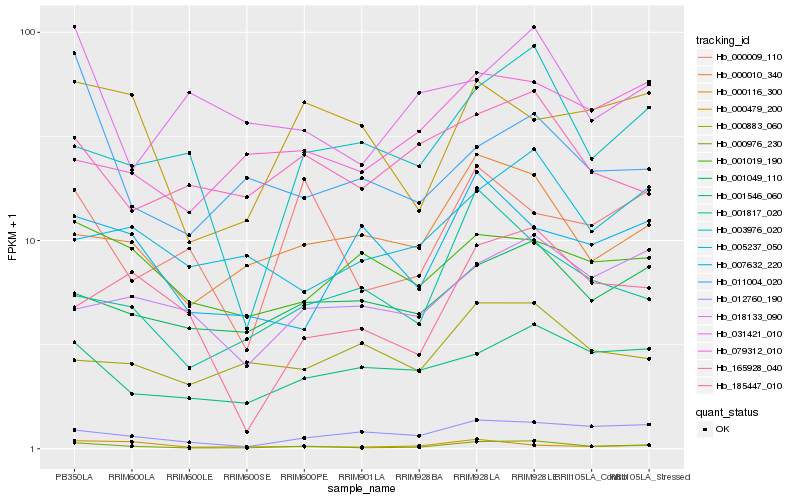
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_230 |
0.0 |
- |
- |
PREDICTED: probable terpene synthase 4 [Jatropha curcas] |
| 2 |
Hb_000010_340 |
0.1804978806 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50280, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001049_110 |
0.1955441327 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_001546_060 |
0.1965400739 |
- |
- |
PREDICTED: uncharacterized protein LOC105133943 isoform X4 [Populus euphratica] |
| 5 |
Hb_012760_190 |
0.1970613459 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 6 |
Hb_003976_020 |
0.1995629609 |
- |
- |
Regulator of ribonuclease activity A, putative [Ricinus communis] |
| 7 |
Hb_001817_020 |
0.2007613133 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 8 |
Hb_005237_050 |
0.2031805656 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105630924 [Jatropha curcas] |
| 9 |
Hb_165928_040 |
0.2060926287 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase component HRD3A [Jatropha curcas] |
| 10 |
Hb_185447_010 |
0.2064200307 |
- |
- |
hypothetical protein JCGZ_13512 [Jatropha curcas] |
| 11 |
Hb_000009_110 |
0.2075595436 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 12 |
Hb_031421_010 |
0.2125042091 |
- |
- |
PREDICTED: copper chaperone for superoxide dismutase, chloroplastic/cytosolic [Jatropha curcas] |
| 13 |
Hb_018133_090 |
0.2163042794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000883_060 |
0.2163712831 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3-like [Jatropha curcas] |
| 15 |
Hb_079312_010 |
0.2183480002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000116_300 |
0.2212733781 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001019_190 |
0.2212942609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007632_220 |
0.2217741568 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 19 |
Hb_000479_200 |
0.2226294052 |
- |
- |
PREDICTED: uncharacterized protein LOC105644128 [Jatropha curcas] |
| 20 |
Hb_011004_020 |
0.2229466249 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |