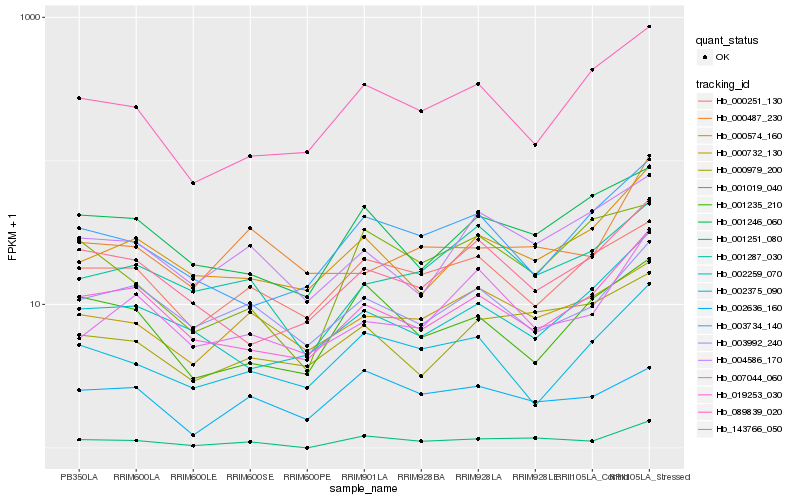
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001251_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_019253_030 |
0.1856304801 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP19-3-like [Citrus sinensis] |
| 3 |
Hb_000574_160 |
0.1922906704 |
- |
- |
PREDICTED: arogenate dehydrogenase 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001019_040 |
0.1942831623 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001246_060 |
0.195513989 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_001287_030 |
0.1961281298 |
- |
- |
30S ribosomal protein S31, mitochondrial precursor, putative [Ricinus communis] |
| 7 |
Hb_000979_200 |
0.2024871409 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 8 |
Hb_089839_020 |
0.2026494925 |
- |
- |
hypothetical protein POPTR_0017s14180g [Populus trichocarpa] |
| 9 |
Hb_003734_140 |
0.2043353377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003992_240 |
0.2045925129 |
- |
- |
- |
| 11 |
Hb_004586_170 |
0.206016495 |
- |
- |
hypothetical protein PRUPE_ppa011196mg [Prunus persica] |
| 12 |
Hb_001235_210 |
0.2067411599 |
- |
- |
PREDICTED: asparagine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 13 |
Hb_000732_130 |
0.2080577756 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_17108 [Jatropha curcas] |
| 14 |
Hb_143766_050 |
0.2093117221 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-5-like [Populus euphratica] |
| 15 |
Hb_007044_060 |
0.2093565569 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_002375_090 |
0.2101563673 |
- |
- |
- |
| 17 |
Hb_000251_130 |
0.2101944476 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 18 |
Hb_000487_230 |
0.2107361015 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 19 |
Hb_002259_070 |
0.2108992027 |
- |
- |
taz protein, putative [Ricinus communis] |
| 20 |
Hb_002636_160 |
0.2114701838 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g13630 [Jatropha curcas] |