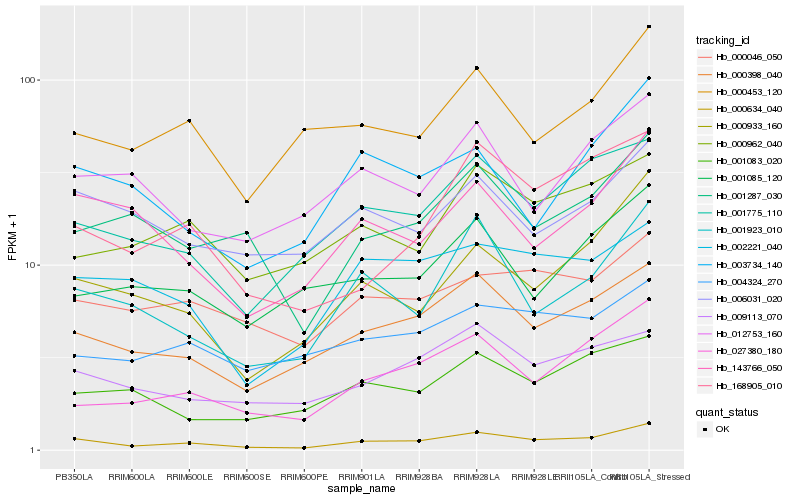
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000634_040 |
0.0 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000398_040 |
0.1039793808 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 3 |
Hb_001775_110 |
0.1201147045 |
- |
- |
PREDICTED: probable ribosome biogenesis protein RLP24 [Jatropha curcas] |
| 4 |
Hb_000933_160 |
0.1263483168 |
- |
- |
PREDICTED: uncharacterized protein LOC105630505 [Jatropha curcas] |
| 5 |
Hb_001923_010 |
0.1268745858 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_027380_180 |
0.1276696664 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 7 |
Hb_009113_070 |
0.133432069 |
- |
- |
hypothetical protein POPTR_0010s07510g [Populus trichocarpa] |
| 8 |
Hb_143766_050 |
0.1361810574 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-5-like [Populus euphratica] |
| 9 |
Hb_168905_010 |
0.1378827318 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-1 isoform X2 [Cicer arietinum] |
| 10 |
Hb_000453_120 |
0.1401785989 |
- |
- |
PREDICTED: proteasome subunit alpha type-3 [Jatropha curcas] |
| 11 |
Hb_001287_030 |
0.140858149 |
- |
- |
30S ribosomal protein S31, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_000046_050 |
0.1423967283 |
- |
- |
PREDICTED: LEC14B homolog [Jatropha curcas] |
| 13 |
Hb_002221_040 |
0.1426693456 |
- |
- |
PREDICTED: uncharacterized protein LOC105639851 [Jatropha curcas] |
| 14 |
Hb_001083_020 |
0.1431335028 |
- |
- |
- |
| 15 |
Hb_003734_140 |
0.1440537462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000962_040 |
0.1458849791 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_006031_020 |
0.1460192282 |
- |
- |
PREDICTED: protein FLX-like 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001085_120 |
0.1475008181 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_012753_160 |
0.1481272169 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 20 |
Hb_004324_270 |
0.14860938 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 [Jatropha curcas] |