| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
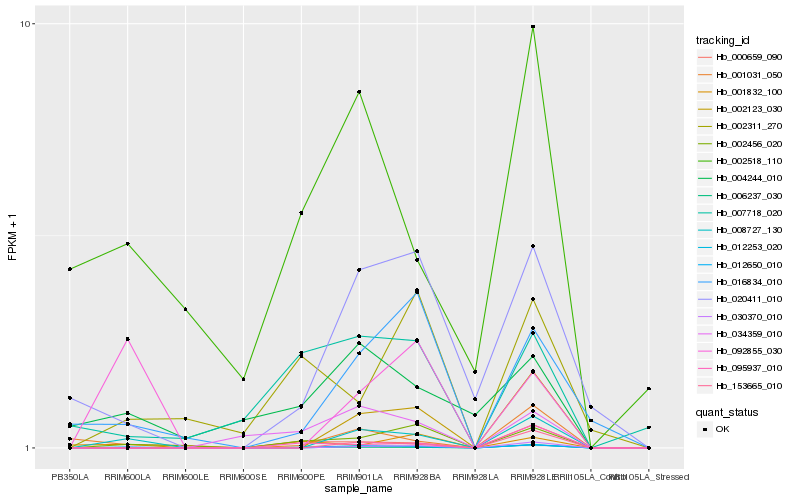
Hb_012650_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 2 |
Hb_008727_130 |
0.1962434021 |
- |
- |
hypothetical protein EUGRSUZ_J01958 [Eucalyptus grandis] |
| 3 |
Hb_002456_020 |
0.235291631 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 4 |
Hb_020411_010 |
0.3193233376 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_016834_010 |
0.3195631339 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 6 |
Hb_012253_020 |
0.3255716977 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_000659_090 |
0.3259104806 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 8 |
Hb_006237_030 |
0.3266926 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_002123_030 |
0.329755076 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 10 |
Hb_092855_030 |
0.3304729649 |
- |
- |
- |
| 11 |
Hb_002518_110 |
0.3310498976 |
- |
- |
1-amino-cyclopropane-1-carboxylic acid oxidase [Manihot esculenta] |
| 12 |
Hb_034359_010 |
0.3362515647 |
- |
- |
- |
| 13 |
Hb_004244_010 |
0.3384447696 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 14 |
Hb_001832_100 |
0.3436572957 |
- |
- |
- |
| 15 |
Hb_001031_050 |
0.3478940079 |
- |
- |
- |
| 16 |
Hb_007718_020 |
0.3496336953 |
- |
- |
PREDICTED: receptor-like protein 12 [Camelina sativa] |
| 17 |
Hb_002311_270 |
0.3544833474 |
- |
- |
PREDICTED: WD repeat-containing protein 43-like [Malus domestica] |
| 18 |
Hb_030370_010 |
0.3560003852 |
- |
- |
PREDICTED: uncharacterized protein LOC104896222 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_095937_010 |
0.3611529686 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 20 |
Hb_153665_010 |
0.3612323175 |
- |
- |
PREDICTED: uncharacterized protein LOC104245945 [Nicotiana sylvestris] |