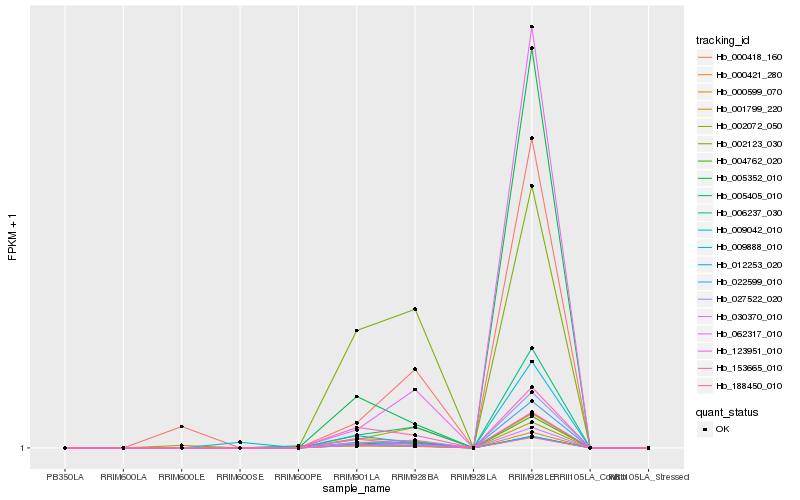
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009042_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_188450_010 |
0.0009260259 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_000599_070 |
0.0204687828 |
- |
- |
PREDICTED: patellin-4-like [Jatropha curcas] |
| 4 |
Hb_153665_010 |
0.0506217166 |
- |
- |
PREDICTED: uncharacterized protein LOC104245945 [Nicotiana sylvestris] |
| 5 |
Hb_123951_010 |
0.0851927371 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_004762_020 |
0.0943010227 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_005405_010 |
0.0983304945 |
- |
- |
- |
| 8 |
Hb_030370_010 |
0.1121895892 |
- |
- |
PREDICTED: uncharacterized protein LOC104896222 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_012253_020 |
0.1415240325 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_002123_030 |
0.1583556439 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 11 |
Hb_022599_010 |
0.1837383848 |
- |
- |
Uncharacterized protein TCM_010507 [Theobroma cacao] |
| 12 |
Hb_005352_010 |
0.1904665807 |
- |
- |
- |
| 13 |
Hb_001799_220 |
0.1941322388 |
transcription factor |
TF Family: AP2 |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000418_160 |
0.21765637 |
- |
- |
- |
| 15 |
Hb_009888_010 |
0.2204561819 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_027522_020 |
0.2225131135 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_062317_010 |
0.2253790329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002072_050 |
0.2289816677 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_000421_280 |
0.2289886646 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 20 |
Hb_006237_030 |
0.2494067015 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |